| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 14:01:37 -0600 |
|---|
| Update Date | 2015-07-14 14:32:03 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | Oxalosuccinic acid |
|---|
| Description | Oxalosuccinate is a substrate for cytoplasmic Isocitrate dehydrogenase and mitochondiral Isocitrate dehydrogenase (mitochondrial). |
|---|
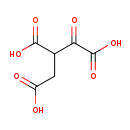
| Structure | |
|---|
| Synonyms: | - Oxalo-succ
- Oxalosuccinate
- Oxalosuccinic acid
|
|---|
| Chemical Formula: | C6H6O7 |
|---|
| Weight: | Average: 190.1076
Monoisotopic: 190.011352546 |
|---|
| InChI Key: | UFSCUAXLTRFIDC-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/C6H6O7/c7-3(8)1-2(5(10)11)4(9)6(12)13/h2H,1H2,(H,7,8)(H,10,11)(H,12,13) |
|---|
| CAS number: | 1948-82-9 |
|---|
| IUPAC Name: | 1-oxopropane-1,2,3-tricarboxylic acid |
|---|
| Traditional IUPAC Name: | oxalosuccinate |
|---|
| SMILES: | OC(=O)CC(C(O)=O)C(=O)C(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as tricarboxylic acids and derivatives. These are carboxylic acids containing exactly three carboxyl groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Tricarboxylic acids and derivatives |
|---|
| Direct Parent | Tricarboxylic acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Tricarboxylic acid or derivatives
- Gamma-keto acid
- Beta-keto acid
- Alpha-keto acid
- Beta-hydroxy ketone
- Keto acid
- 1,3-dicarbonyl compound
- Alpha-hydroxy ketone
- Ketone
- Carboxylic acid
- Organic oxide
- Carbonyl group
- Organooxygen compound
- Hydrocarbon derivative
- Organic oxygen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -3 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | |
|---|
| KEGG Pathways: | - Citrate cycle (TCA cycle) ec00020
- Glutathione metabolism ec00480
- Metabolic pathways eco01100
- Microbial metabolism in diverse environments ec01120
- Reductive carboxylate cycle (CO2 fixation) ec00720
|
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0005-6900000000-05e6953cf97abc6dd674 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-00rf-9137000000-9567484c80debff89c0e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00dj-1900000000-ed9f625da4af156fd85a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-006t-8900000000-b245ec6d4dca3949083d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-006t-9300000000-00b8a65afa1e83c61b5a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000b-0900000000-a6734cca6590d164a554 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-3900000000-c30d22d22f12460fcc5c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00dj-9300000000-a3b40df9f3852aa0e3c8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0fi0-0900000000-de098f1139c0f6fbd19d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0092-9800000000-03bc5902024a449a2d18 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00di-9000000000-ef352a8e83bf22a4b124 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0f6t-3900000000-654a5517d3a1a65425e3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0f6t-7900000000-7b0190686602c20ebac6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0fk9-9500000000-e5f05497c77c1ef44cf0 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
|
|---|
| Synthesis Reference: | Takeda, Isao; Iguchi, Takashi. Fermentative production of oxalosuccinic acid from hydrocarbons. Jpn. Tokkyo Koho (1972), 3 pp. |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|