| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 14:00:42 -0600 |
|---|
| Update Date | 2015-06-03 15:54:33 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | 3-Oxotetradecanoyl-CoA |
|---|
| Description | 3-Oxotetradecanoyl-CoA is a product of the peroxisomal beta oxidation of hexadenoic acid by the enzyme acyl-CoA oxidase which results in long-chain 3-oxoacyl-CoA-esters. (PMID: 7548202) |
|---|
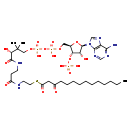
| Structure | |
|---|
| Synonyms: | - 3-Oxomyristoyl-CoA
- 3-Oxomyristoyl-Coenzyme A
- 3-Oxotetradecanoyl-CoA
- 5'-{3-[(3R)-3-Hydroxy-2,2-dimethyl-4-oxo-4-{[3-oxo-3-({2-[(3-oxotetradecanoyl)sulfanyl]ethyl}amino)propyl]amino}butyl] dihydrogen diphosphate}3'-phosphoadenosine
- 5'-{3-[(3R)-3-hydroxy-2,2-dimethyl-4-oxo-4-{[3-oxo-3-({2-[(3-oxotetradecanoyl)sulfanyl]ethyl}amino)propyl]amino}butyl] dihydrogen diphosphoric acid}3'-phosphoadenosine
- 5'-{3-[(3R)-3-Hydroxy-2,2-dimethyl-4-oxo-4-{[3-oxo-3-({2-[(3-oxotetradecanoyl)sulphanyl]ethyl}amino)propyl]amino}butyl] dihydrogen diphosphate}3'-phosphoadenosine
- 5'-{3-[(3R)-3-hydroxy-2,2-dimethyl-4-oxo-4-{[3-oxo-3-({2-[(3-oxotetradecanoyl)sulphanyl]ethyl}amino)propyl]amino}butyl] dihydrogen diphosphoric acid}3'-phosphoadenosine
- S-(3-oxotetradecanoate
- S-(3-Oxotetradecanoate)
- S-(3-Oxotetradecanoate) CoA
- S-(3-Oxotetradecanoate) Coenzyme A
- S-(3-oxotetradecanoic acid
- S-(3-Oxotetradecanoic acid)
- S-(3-Oxotetradecanoic acid) CoA
- S-(3-Oxotetradecanoic acid) coenzyme A
|
|---|
| Chemical Formula: | C35H60N7O18P3S |
|---|
| Weight: | Average: 991.873
Monoisotopic: 991.292838377 |
|---|
| InChI Key: | IQNFBGHLIVBNOU-QSGBVPJFSA-N |
|---|
| InChI: | InChI=1S/C35H60N7O18P3S/c1-4-5-6-7-8-9-10-11-12-13-23(43)18-26(45)64-17-16-37-25(44)14-15-38-33(48)30(47)35(2,3)20-57-63(54,55)60-62(52,53)56-19-24-29(59-61(49,50)51)28(46)34(58-24)42-22-41-27-31(36)39-21-40-32(27)42/h21-22,24,28-30,34,46-47H,4-20H2,1-3H3,(H,37,44)(H,38,48)(H,52,53)(H,54,55)(H2,36,39,40)(H2,49,50,51)/t24-,28-,29-,30+,34-/m1/s1 |
|---|
| CAS number: | 122364-86-7 |
|---|
| IUPAC Name: | {[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-2-({[hydroxy({hydroxy[(3R)-3-hydroxy-2,2-dimethyl-3-{[2-({2-[(3-oxotetradecanoyl)sulfanyl]ethyl}carbamoyl)ethyl]carbamoyl}propoxy]phosphoryl}oxy)phosphoryl]oxy}methyl)oxolan-3-yl]oxy}phosphonic acid |
|---|
| Traditional IUPAC Name: | 3-oxotetradecanoyl-coa |
|---|
| SMILES: | CCCCCCCCCCCC(=O)CC(=O)SCCNC(=O)CCNC(=O)[C@H](O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C(N)N=CN=C12 |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as long-chain 3-oxoacyl coas. These are organic compounds containing a coenzyme A derivative, which is 3-oxo acylated long aliphatic chain of 13 to 21 carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acyl thioesters |
|---|
| Direct Parent | Long-chain 3-oxoacyl CoAs |
|---|
| Alternative Parents | |
|---|
| Substituents | - Coenzyme a or derivatives
- Purine ribonucleoside 3',5'-bisphosphate
- Purine ribonucleoside bisphosphate
- Purine ribonucleoside diphosphate
- Ribonucleoside 3'-phosphate
- Pentose phosphate
- Pentose-5-phosphate
- Beta amino acid or derivatives
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Monosaccharide phosphate
- Organic pyrophosphate
- Pentose monosaccharide
- Imidazopyrimidine
- Purine
- Monoalkyl phosphate
- Aminopyrimidine
- 1,3-dicarbonyl compound
- Imidolactam
- Monosaccharide
- N-acyl-amine
- N-substituted imidazole
- Organic phosphoric acid derivative
- Pyrimidine
- Alkyl phosphate
- Fatty amide
- Phosphoric acid ester
- Tetrahydrofuran
- Imidazole
- Azole
- Heteroaromatic compound
- Carbothioic s-ester
- Secondary alcohol
- Ketone
- Thiocarboxylic acid ester
- Carboxamide group
- Secondary carboxylic acid amide
- Amino acid or derivatives
- Sulfenyl compound
- Thiocarboxylic acid or derivatives
- Organoheterocyclic compound
- Azacycle
- Oxacycle
- Carboxylic acid derivative
- Organosulfur compound
- Organic oxygen compound
- Hydrocarbon derivative
- Carbonyl group
- Organic nitrogen compound
- Primary amine
- Organopnictogen compound
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Amine
- Alcohol
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -4 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | |
|---|
| KEGG Pathways: | |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-1902100203-37e47fa131f279d1ccdb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0922300000-89842e38a36b7c108b45 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-1900101000-4f1b2969f929942e67b3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00al-3951231305-e64e775b7655f9988bfe | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-3921200001-fab03116b2cfb433bbae | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-057i-6900100000-457e07aa65d0f83247fb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0000000009-342646973b6a3f3d5bf3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00g3-4200201329-23aa6cf502f4d804fa11 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-02jc-2101301309-a62c66b8cd13c607e063 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-006x-0000000009-01e6acdaa109155ae7aa | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0600100189-f65f6ff48a5a3738933c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-0001900000-d1155a9305d5c03af39a | View in MoNA |
|---|
| MS | Mass Spectrum (Electron Ionization) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- Kishore NS, Wood DC, Mehta PP, Wade AC, Lu T, Gokel GW, Gordon JI: Comparison of the acyl chain specificities of human myristoyl-CoA synthetase and human myristoyl-CoA:protein N-myristoyltransferase. J Biol Chem. 1993 Mar 5;268(7):4889-902. Pubmed: 8444867
- Nagi MN, Cook L, Suneja SK, Osei P, Cinti DL: Spectrophotometric assay for the condensing enzyme activity of the microsomal fatty acid chain elongation system. Anal Biochem. 1989 Jun;179(2):251-61. Pubmed: 2774174
- Sleboda, J., Pourfarzam, M., Bartlett, K., Osmundsen, H. (1995). "Effects of added l-carnitine, acetyl-CoA and CoA on peroxisomal beta-oxidation of [U-14C]hexadecanoate by isolated peroxisomal fractions." Biochim Biophys Acta 1258:309-318. Pubmed: 7548202
- Watmough NJ, Turnbull DM, Sherratt HS, Bartlett K: Measurement of the acyl-CoA intermediates of beta-oxidation by h.p.l.c. with on-line radiochemical and photodiode-array detection. Application to the study of [U-14C]hexadecanoate oxidation by intact rat liver mitochondria. Biochem J. 1989 Aug 15;262(1):261-9. Pubmed: 2818568
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|