Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
Myo-inositol hexakisphosphate (M2MDB000507)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-05-31 13:59:25 -0600 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-09-17 15:41:17 -0600 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Myo-inositol hexakisphosphate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Myo-Inositol hexakisphosphate is an intermediate in inositol phosphate metabolism. It can be generated from D-myo-Inositol 1,3,4,5,6-pentakisphosphate via the enzyme inositol-pentakisphosphate 2-kinase (EC:2.7.1.158). Myo-Inositol hexakisphosphate is also known as phytic acid. It can be used clinically as a complexing agent for removal of traces of heavy metal ions. It acts also as a hypocalcemic agent. Phytic acid is a strong chelator of important minerals such as calcium, magnesium, iron and zinc. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
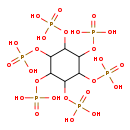
| Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C6H18O24P6 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 660.0353 Monoisotopic: 659.861370576 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | IMQLKJBTEOYOSI-UHFFFAOYSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C6H18O24P6/c7-31(8,9)25-1-2(26-32(10,11)12)4(28-34(16,17)18)6(30-36(22,23)24)5(29-35(19,20)21)3(1)27-33(13,14)15/h1-6H,(H2,7,8,9)(H2,10,11,12)(H2,13,14,15)(H2,16,17,18)(H2,19,20,21)(H2,22,23,24) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 83-86-3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | {[2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl]oxy}phosphonic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | phytic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | OP(O)(=O)OC1C(OP(O)(O)=O)C(OP(O)(O)=O)C(OP(O)(O)=O)C(OP(O)(O)=O)C1OP(O)(O)=O | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as inositol phosphates. Inositol phosphates are compounds containing a phosphate group attached to an inositol (or cyclohexanehexol) moiety. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic oxygen compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Organooxygen compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Alcohols and polyols | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Inositol phosphates | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic homomonocyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -12 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | < 25 °C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | 6 Water + Myo-inositol hexakisphosphate > Inositol +6 Phosphate Myo-inositol hexakisphosphate + Water <> 1-Myo-inositol 1,2,3,4,5-pentakisphosphate + Phosphate Myo-inositol hexakisphosphate + Water > 1-Myo-inositol 1,2,3,4,5-pentakisphosphate + Phosphate Myo-inositol hexakisphosphate + Water > D-<i>myo</i>-inositol (1,2,4,5,6)-pentakisphosphate + Phosphate Myo-inositol hexakisphosphate + Water > 1D-myo-inositol 1,2,3,5,6-pentakisphosphate + Inorganic phosphate Myo-inositol hexakisphosphate + Water + Myo-inositol hexakisphosphate > 1-Myo-inositol 1,2,3,4,5-pentakisphosphate + Phosphate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Mitsuhashi Naoto; Ohnishi Miwa; Sekiguchi Yoko; Kwon Yong-Uk; Chang Young-Tae; Chung Sung-Kee; Inoue Yoshinori; Reid Robert J; Yagisawa Hitoshi; Mimura Tetsuro Phytic acid synthesis and vacuolar accumulation in suspension-cultured cells of Catharanthus r | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in acid phosphatase activity
- Specific function:
- A phosphate monoester + H(2)O = an alcohol + phosphate
- Gene Name:
- appA
- Uniprot ID:
- P07102
- Molecular weight:
- 47056
Reactions
| A phosphate monoester + H(2)O = an alcohol + phosphate. |
| Myo-inositol hexakisphosphate + H(2)O = 1D-myo-inositol 1,2,3,5,6-pentakisphosphate + phosphate. |