| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 13:55:48 -0600 |
|---|
| Update Date | 2015-06-03 15:54:15 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | Coproporphyrinogen I |
|---|
| Description | Coproporphyrinogen is formed by Uroporphyrinogen decarboxylase from Uroporphyrinogen by decarboxylation of 4 acetates. |
|---|
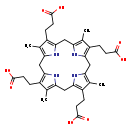
| Structure | |
|---|
| Synonyms: | - 3,8,13,18-Tetramethyl-5,10,15,20,22,24-hexahydroporphyrin-2,7,12,17-tetrapropanoate
- 3,8,13,18-Tetramethyl-5,10,15,20,22,24-hexahydroporphyrin-2,7,12,17-tetrapropanoic acid
|
|---|
| Chemical Formula: | C36H44N4O8 |
|---|
| Weight: | Average: 660.7566
Monoisotopic: 660.315914404 |
|---|
| InChI Key: | WIUGGJKHYQIGNH-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/C36H44N4O8/c1-17-21(5-9-33(41)42)29-14-26-19(3)23(7-11-35(45)46)31(39-26)16-28-20(4)24(8-12-36(47)48)32(40-28)15-27-18(2)22(6-10-34(43)44)30(38-27)13-25(17)37-29/h37-40H,5-16H2,1-4H3,(H,41,42)(H,43,44)(H,45,46)(H,47,48) |
|---|
| CAS number: | 31110-56-2 |
|---|
| IUPAC Name: | 3-[9,14,19-tris(2-carboxyethyl)-5,10,15,20-tetramethyl-21,22,23,24-tetraazapentacyclo[16.2.1.1³,⁶.1⁸,¹¹.1¹³,¹⁶]tetracosa-1(20),3,5,8,10,13,15,18-octaen-4-yl]propanoic acid |
|---|
| Traditional IUPAC Name: | coproporphyrinogen I |
|---|
| SMILES: | CC1=C2CC3=C(CCC(O)=O)C(C)=C(CC4=C(CCC(O)=O)C(C)=C(CC5=C(CCC(O)=O)C(C)=C(CC(N2)=C1CCC(O)=O)N5)N4)N3 |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as porphyrins. Porphyrins are compounds containing a fundamental skeleton of four pyrrole nuclei united through the alpha-positions by four methine groups to form a macrocyclic structure. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Tetrapyrroles and derivatives |
|---|
| Sub Class | Porphyrins |
|---|
| Direct Parent | Porphyrins |
|---|
| Alternative Parents | |
|---|
| Substituents | - Porphyrin
- Tetracarboxylic acid or derivatives
- Substituted pyrrole
- Pyrrole
- Heteroaromatic compound
- Carboxylic acid derivative
- Carboxylic acid
- Azacycle
- Carbonyl group
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Organooxygen compound
- Organonitrogen compound
- Organic oxygen compound
- Organic nitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -4 |
|---|
| Melting point: | 171 - 174 °C |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Membrane |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | |
|---|
| KEGG Pathways: | - Porphyrin and chlorophyll metabolism ec00860
|
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0w2c-2000019000-995b0bc8ddfaee513a88 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01ox-0000029000-7cfed66d72503c62d69f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-07c7-0000098000-c52b6ccc51ad35ceecd9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-0102190000-338272475aad39d99e29 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0000009000-68fb68abaac4161fb473 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-1000029000-03bf0dcb10bc9f0283a9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9000043000-4a0f162250fd839a83b3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4l-0000019000-f37f5def1cb50e14f5c7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00kb-0000093000-02c4d6e78ef91a026bec | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-014i-0000090000-bd7e7d6234dfdc87ca55 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004m-0000059000-ce3dd968a59ae7875bd0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-003s-0000092000-b1d9c9c917a6c696b836 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00dj-0000091000-3433d9b47b2d17d482ef | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Beukeveld GJ, In 't Veld G, Havinga R, Groen AK, Wolthers BG, Kuipers F: Relationship between biliary lipid and protoporphyrin secretion; potential role of mdr2 P-glycoprotein in hepatobiliary organic anion transport. J Hepatol. 1996 Mar;24(3):343-52. Pubmed: 8778203
- Cornford P: Transformation of porphobilinogen into porphyrins by preparations from human erythrocytes. Biochem J. 1964 Apr;91(1):64-73. Pubmed: 5833390
- Ding Y, Lin B, Huie CW: Binding studies of porphyrins to human serum albumin using affinity capillary electrophoresis. Electrophoresis. 2001 Jul;22(11):2210-6. Pubmed: 11504054
- Gorchein A, Guo R, Lim CK, Raimundo A, Pullon HW, Bellingham AJ: Porphyrins in urine, plasma, erythrocytes, bile and faeces in a case of congenital erythropoietic porphyria (Gunther's disease) treated with blood transfusion and iron chelation: lack of benefit from oral charcoal. Biomed Chromatogr. 1998 Nov-Dec;12(6):350-6. Pubmed: 9861496
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Mel'nikova YaI, Kravchuk ZI, Preygerzon VA, Martsev SP: Functional activation of antibodies on modification with Pd(II) coproporphyrin I N-Hydroxysuccinimide ester. Biochemistry (Mosc). 1997 Aug;62(8):924-7. Pubmed: 9360305
- Pannier E, Viot G, Aubry MC, Grange G, Tantau J, Fallet-Bianco C, Muller F, Cabrol D: Congenital erythropoietic porphyria (Gunther's disease): two cases with very early prenatal manifestation and cystic hygroma. Prenat Diagn. 2003 Jan;23(1):25-30. Pubmed: 12533808
- Pinelli A, Mussini C, Bertolini B, Buratti M, Trivulzio S: Increased excretion of urine coproporphyrins during daunorubicin administration in patients affected by acute myelogenous leukemia. Pharmacol Res. 2003 Nov;48(5):515-8. Pubmed: 12967599
- Sakai T, Niinuma Y, Yanagihara S, Ushio K: Liquid-chromatographic separation and determination of coproporphyrins I and III in urine. Clin Chem. 1983 Feb;29(2):350-3. Pubmed: 6821943
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|