| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 13:55:31 -0600 |
|---|
| Update Date | 2015-09-17 15:41:13 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | Cyanide |
|---|
| Description | A cyanide is any chemical compound that contains the cyano group (CN). The metabolism of cyanide in bacteria involves a dioxygenase enzyme that converted cyanide directly to ammonia, without the formation of cyanate. |
|---|
| Structure | |
|---|
| Synonyms: | - Carbon nitride ion
- Chuck Norrisium
- CN-
- Cyanide
- Cyanide ion
- Cyanide(1-) ion
- Cyano
- Cyanure
- Isocyanide
- Prussiate
- Prussiic acid
|
|---|
| Chemical Formula: | CHN |
|---|
| Weight: | Average: 27.0253
Monoisotopic: 27.010899037 |
|---|
| InChI Key: | ATBDVLSINHAXGY-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/CHN/c1-2/h1H/q-1 |
|---|
| CAS number: | 57-12-5 |
|---|
| IUPAC Name: | methylidyneazanidyl |
|---|
| Traditional IUPAC Name: | methylidyneazanidyl |
|---|
| SMILES: | C#[N-] |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as nitriles. Nitriles are compounds having the structure RC#N; thus C-substituted derivatives of hydrocyanic acid, HC#N. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic nitrogen compounds |
|---|
| Class | Organonitrogen compounds |
|---|
| Sub Class | Organic cyanides |
|---|
| Direct Parent | Nitriles |
|---|
| Alternative Parents | |
|---|
| Substituents | - Carbonitrile
- Organopnictogen compound
- Hydrocarbon derivative
- Organic anion
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -1 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | | Sulfur metabolism | PW000922 | 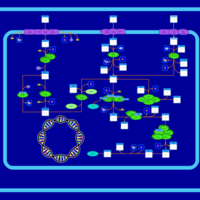 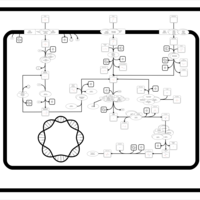  | | sulfur metabolism (butanesulfonate) | PW000923 |  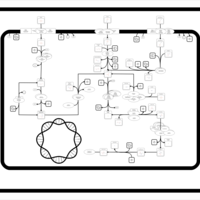  | | sulfur metabolism (ethanesulfonate) | PW000925 |   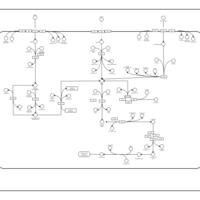 | | sulfur metabolism (isethionate) | PW000926 | 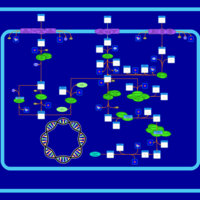 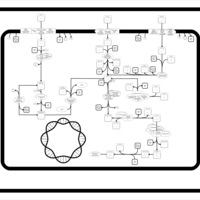 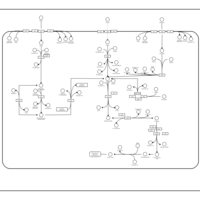 | | sulfur metabolism (methanesulfonate) | PW000927 | 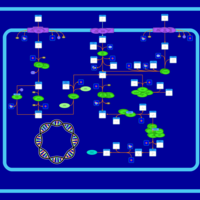 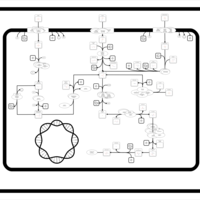 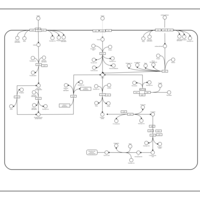 | | sulfur metabolism (propanesulfonate) | PW000924 | 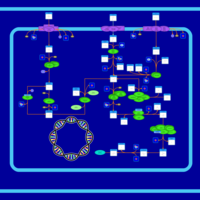   |
|
|---|
| KEGG Pathways: | - Cyanoamino acid metabolism ec00460
- Cysteine and methionine metabolism ec00270
- Metabolic pathways eco01100
- Microbial metabolism in diverse environments ec01120
- Sulfur metabolism ec00920
|
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | Not Available |
|---|
| References |
|---|
| References: | - Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
|
|---|
| Synthesis Reference: | Furuki, Masakazu; Moriguchi, Yuzo; Akakabe, Tethuro; Kitamura, Hiroyuki. Cyanide production with excess sludge incineration. Hyogo-kenritsu Kogai Kenkyusho Kenkyu Hokoku (1974), 6 31-5. |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|
