Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
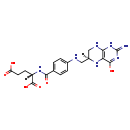
Tetrahydrofolic acid (M2MDB000424)
Enzymes
- General function:
- Involved in hydroxymethyl-, formyl- and related transferase activity
- Specific function:
- 10-formyltetrahydrofolate + N(1)-(5-phospho-D- ribosyl)glycinamide = tetrahydrofolate + N(2)-formyl-N(1)-(5- phospho-D-ribosyl)glycinamide
- Gene Name:
- purN
- Uniprot ID:
- P08179
- Molecular weight:
- 23238
Reactions
| 10-formyltetrahydrofolate + N(1)-(5-phospho-D-ribosyl)glycinamide = tetrahydrofolate + N(2)-formyl-N(1)-(5-phospho-D-ribosyl)glycinamide. |
- General function:
- Involved in tetrahydrofolylpolyglutamate synthase activity
- Specific function:
- Conversion of folates to polyglutamate derivatives
- Gene Name:
- folC
- Uniprot ID:
- P08192
- Molecular weight:
- 45405
Reactions
| ATP + tetrahydropteroyl-(gamma-Glu)(n) + L-glutamate = ADP + phosphate + tetrahydropteroyl-(gamma-Glu)(n+1). |
| ATP + 7,8-dihydropteroate + L-glutamate = ADP + phosphate + 7,8-dihydropteroylglutamate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Interconversion of serine and glycine
- Gene Name:
- glyA
- Uniprot ID:
- P0A825
- Molecular weight:
- 45316
Reactions
| 5,10-methylenetetrahydrofolate + glycine + H(2)O = tetrahydrofolate + L-serine. |
- General function:
- Involved in oxidoreductase activity
- Specific function:
- Lipoamide dehydrogenase is a component of the glycine cleavage system as well as of the alpha-ketoacid dehydrogenase complexes
- Gene Name:
- lpdA
- Uniprot ID:
- P0A9P0
- Molecular weight:
- 50688
Reactions
| Protein N(6)-(dihydrolipoyl)lysine + NAD(+) = protein N(6)-(lipoyl)lysine + NADH. |
- General function:
- Involved in dihydrofolate reductase activity
- Specific function:
- Key enzyme in folate metabolism. Catalyzes an essential reaction for de novo glycine and purine synthesis, and for DNA precursor synthesis
- Gene Name:
- folA
- Uniprot ID:
- P0ABQ4
- Molecular weight:
- 17999
Reactions
| 5,6,7,8-tetrahydrofolate + NADP(+) = 7,8-dihydrofolate + NADPH. |
- General function:
- Involved in oxidoreductase activity
- Specific function:
- Catalyzes the reduction of dihydrofolate to tetrahydrofolate
- Gene Name:
- folM
- Uniprot ID:
- P0AFS3
- Molecular weight:
- 26348
Reactions
| 5,6,7,8-tetrahydrofolate + NADP(+) = 7,8-dihydrofolate + NADPH. |
- General function:
- Involved in methionine synthase activity
- Specific function:
- Catalyzes the transfer of a methyl group from methyl- cobalamin to homocysteine, yielding enzyme-bound cob(I)alamin and methionine. Subsequently, remethylates the cofactor using methyltetrahydrofolate
- Gene Name:
- metH
- Uniprot ID:
- P13009
- Molecular weight:
- 135996
Reactions
| 5-methyltetrahydrofolate + L-homocysteine = tetrahydrofolate + L-methionine. |
- General function:
- Involved in IMP cyclohydrolase activity
- Specific function:
- 10-formyltetrahydrofolate + 5-amino-1-(5- phospho-D-ribosyl)imidazole-4-carboxamide = tetrahydrofolate + 5- formamido-1-(5-phospho-D-ribosyl)imidazole-4-carboxamide
- Gene Name:
- purH
- Uniprot ID:
- P15639
- Molecular weight:
- 57329
Reactions
| 10-formyltetrahydrofolate + 5-amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxamide = tetrahydrofolate + 5-formamido-1-(5-phospho-D-ribosyl)imidazole-4-carboxamide. |
| IMP + H(2)O = 5-formamido-1-(5-phospho-D-ribosyl)imidazole-4-carboxamide. |
- General function:
- Involved in methionyl-tRNA formyltransferase activity
- Specific function:
- Modifies the free amino group of the aminoacyl moiety of methionyl-tRNA(fMet). The formyl group appears to play a dual role in the initiator identity of N-formylmethionyl-tRNA by:(I) promoting its recognition by IF2 and (II) impairing its binding to EFTu-GTP
- Gene Name:
- fmt
- Uniprot ID:
- P23882
- Molecular weight:
- 34168
Reactions
| 10-formyltetrahydrofolate + L-methionyl-tRNA(fMet) + H(2)O = tetrahydrofolate + N-formylmethionyl-tRNA(fMet). |
- General function:
- Involved in 5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase activity
- Specific function:
- Catalyzes the transfer of a methyl group from 5- methyltetrahydrofolate to homocysteine resulting in methionine formation
- Gene Name:
- metE
- Uniprot ID:
- P25665
- Molecular weight:
- 84673
Reactions
| 5-methyltetrahydropteroyltri-L-glutamate + L-homocysteine = tetrahydropteroyltri-L-glutamate + L-methionine. |
- General function:
- Involved in aminomethyltransferase activity
- Specific function:
- The glycine cleavage system catalyzes the degradation of glycine
- Gene Name:
- gcvT
- Uniprot ID:
- P27248
- Molecular weight:
- 40146
Reactions
| [Protein]-S(8)-aminomethyldihydrolipoyllysine + tetrahydrofolate = [protein]-dihydrolipoyllysine + 5,10-methylenetetrahydrofolate + NH(3). |
- General function:
- Involved in 3-methyl-2-oxobutanoate hydroxymethyltransferase activity
- Specific function:
- Catalyzes the reversible reaction in which hydroxymethyl group from 5,10-methylenetetrahydrofolate is tranferred onto alpha-ketoisovalerate to form ketopantoate
- Gene Name:
- panB
- Uniprot ID:
- P31057
- Molecular weight:
- 28237
Reactions
| 5,10-methylenetetrahydrofolate + 3-methyl-2-oxobutanoate + H(2)O = tetrahydrofolate + 2-dehydropantoate. |
- General function:
- Involved in glycine dehydrogenase (decarboxylating) activity
- Specific function:
- The glycine cleavage system catalyzes the degradation of glycine. The P protein binds the alpha-amino group of glycine through its pyridoxal phosphate cofactor; CO(2) is released and the remaining methylamine moiety is then transferred to the lipoamide cofactor of the H protein
- Gene Name:
- gcvP
- Uniprot ID:
- P33195
- Molecular weight:
- 104376
Reactions
| Glycine + H-protein-lipoyllysine = H-protein-S-aminomethyldihydrolipoyllysine + CO(2). |
- General function:
- Involved in ATP binding
- Specific function:
- Catalyzes two reactions:the first one is the production of beta-formyl glycinamide ribonucleotide (GAR) from formate, ATP and beta GAR; the second, a side reaction, is the production of acetyl phosphate and ADP from acetate and ATP
- Gene Name:
- purT
- Uniprot ID:
- P33221
- Molecular weight:
- 42433
Reactions
| Formate + ATP + 5'-phospho-ribosylglycinamide = 5'-phosphoribosyl-N-formylglycinamide + ADP + diphosphate. |
- General function:
- Involved in amino acid binding
- Specific function:
- Produces formate from formyl-tetrahydrofolate. Provides the major source of formate for the purT-dependent synthesis of 5'-phosphoribosyl-N-formylglycinamide (FGAR) during aerobic growth. Has a role in regulating the one-carbon pool
- Gene Name:
- purU
- Uniprot ID:
- P37051
- Molecular weight:
- 31934
Reactions
| 10-formyltetrahydrofolate + H(2)O = formate + tetrahydrofolate. |
- General function:
- Involved in hydroxymethyl-, formyl- and related transferase activity
- Specific function:
- Bifunctional enzyme that catalyzes the oxidative decarboxylation of UDP-glucuronic acid (UDP-GlcUA) to UDP-4-keto- arabinose (UDP-Ara4O) and the addition of a formyl group to UDP-4- amino-4-deoxy-L-arabinose (UDP-L-Ara4N) to form UDP-L-4-formamido- arabinose (UDP-L-Ara4FN). The modified arabinose is attached to lipid A and is required for resistance to polymyxin and cationic antimicrobial peptides
- Gene Name:
- arnA
- Uniprot ID:
- P77398
- Molecular weight:
- 74288
Reactions
| UDP-alpha-D-glucuronate + NAD(+) = UDP-beta-L-threo-pentapyranos-4-ulose + CO(2) + NADH. |
| 10-formyltetrahydrofolate + UDP-4-amino-4-deoxy-beta-L-arabinose = 5,6,7,8-tetrahydrofolate + UDP-4-deoxy-4-formamido-beta-L-arabinose. |
- General function:
- Amino acid transport and metabolism
- Specific function:
- The glycine cleavage system catalyzes the degradation of glycine. The H protein shuttles the methylamine group of glycine from the P protein to the T protein
- Gene Name:
- gcvH
- Uniprot ID:
- P0A6T9
- Molecular weight:
- 13811