Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
Xanthylic acid (M2MDB000419)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-05-31 13:54:28 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 15:54:12 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Xanthylic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Xanthylic acid is an important metabolic intermediate in the purine metabolism, and is a product or substrate of the enzymes inosine monophosphate dehydrogenase (EC 1.1.1.205), hypoxanthine phosphoribosyltransferase (EC 2.4.2.8), xanthine phosphoribosyltransferase (EC 2.4.2.22), 5'-ribonucleotide phosphohydrolase (EC 3.1.3.5), Ap4A hydrolase (EC 3.6.1.17), nucleoside-triphosphate diphosphatase (EC 3.6.1.19), phosphoribosylamine-glycine ligase (EC 6.3.4.1), and glutamine amidotransferase (EC 6.3.5.2). (KEGG) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
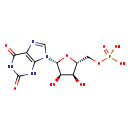
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C10H13N4O9P | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 364.2054 Monoisotopic: 364.042014546 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | DCTLYFZHFGENCW-UUOKFMHZSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C10H13N4O9P/c15-5-3(1-22-24(19,20)21)23-9(6(5)16)14-2-11-4-7(14)12-10(18)13-8(4)17/h2-3,5-6,9,15-16H,1H2,(H2,19,20,21)(H2,12,13,17,18)/t3-,5-,6-,9-/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 523-98-8 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | {[(2R,3S,4R,5R)-5-(2,6-dioxo-2,3,6,9-tetrahydro-1H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}phosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | xanthosine monophosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | O[C@@H]1[C@@H](COP(O)(O)=O)O[C@H]([C@@H]1O)N1C=NC2=C1NC(=O)NC2=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as purine ribonucleoside monophosphates. These are nucleotides consisting of a purine base linked to a ribose to which one monophosphate group is attached. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Nucleosides, nucleotides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Purine nucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Purine ribonucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Purine ribonucleoside monophosphates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Water + Xanthylic acid > Phosphate + Xanthosine Phosphoribosyl pyrophosphate + Xanthine <> Pyrophosphate + Xanthylic acid Adenosine triphosphate + L-Glutamine + Water + Xanthylic acid > Adenosine monophosphate + L-Glutamate + Guanosine monophosphate +2 Hydrogen ion + Pyrophosphate Water + Inosinic acid + NAD <> Hydrogen ion + NADH + Xanthylic acid Water + Xanthosine 5-triphosphate > Hydrogen ion + Pyrophosphate + Xanthylic acid Adenosine triphosphate + Xanthylic acid + Ammonia <> Adenosine monophosphate + Pyrophosphate + Guanosine monophosphate Adenosine triphosphate + Xanthylic acid + L-Glutamine + Water <> Adenosine monophosphate + Pyrophosphate + Guanosine monophosphate + L-Glutamate Xanthylic acid + Pyrophosphate <> Xanthine + Phosphoribosyl pyrophosphate Xanthylic acid + Water <> Xanthosine + Phosphate Xanthosine 5-triphosphate + Water <> Xanthylic acid + Pyrophosphate Water + L-Glutamine + Xanthylic acid + Adenosine triphosphate > Hydrogen ion + L-Glutamate + Guanosine monophosphate + Pyrophosphate + Adenosine monophosphate Adenosine triphosphate + Xanthylic acid + Ammonia > Hydrogen ion + Adenosine monophosphate + Pyrophosphate + Guanosine monophosphate Water + NAD + Inosinic acid > Hydrogen ion + NADH + Xanthylic acid Xanthylic acid + Pyrophosphate < Xanthine + Phosphoribosyl pyrophosphate Adenosine triphosphate + Xanthylic acid + L-Glutamine + Water > Adenosine monophosphate + Pyrophosphate + Guanosine monophosphate + L-Glutamate Inosinic acid + NAD + Water > Xanthylic acid + NADH Xanthylic acid + Pyrophosphate > Phosphoribosyl pyrophosphate + Xanthine Xanthosine 5-triphosphate + Water + 2'-Deoxyinosine triphosphate <> Xanthylic acid + Pyrophosphate + DIMP Adenosine triphosphate + Xanthylic acid + L-Glutamine + Water + Ammonia <> Adenosine monophosphate + Pyrophosphate + Guanosine monophosphate + L-Glutamate Xanthylic acid + Adenosine triphosphate + L-Glutamine + Water > Adenosine monophosphate + Pyrophosphate + L-Glutamic acid +2 Hydrogen ion + Guanosine monophosphate + L-Glutamate Xanthine + Phosphoribosyl pyrophosphate > Xanthylic acid + Pyrophosphate Adenosine triphosphate + L-Glutamine + Water + Xanthylic acid > Adenosine monophosphate + L-Glutamate + Guanosine monophosphate +2 Hydrogen ion + Pyrophosphate Water + Inosinic acid + NAD <> Hydrogen ion + NADH + Xanthylic acid Adenosine triphosphate + L-Glutamine + Water + Xanthylic acid > Adenosine monophosphate + L-Glutamate + Guanosine monophosphate +2 Hydrogen ion + Pyrophosphate Water + Inosinic acid + NAD <> Hydrogen ion + NADH + Xanthylic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Hattori, Kyoji; Kawahara, Shin; Hagiwara, Takeshige. 5'-Xanthylic acid. Jpn. Kokai Tokkyo Koho (1985), 3 pp. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in GMP synthase (glutamine-hydrolyzing) activity
- Specific function:
- Catalyzes the synthesis of GMP from XMP
- Gene Name:
- guaA
- Uniprot ID:
- P04079
- Molecular weight:
- 58679
Reactions
| ATP + xanthosine 5'-phosphate + L-glutamine + H(2)O = AMP + diphosphate + GMP + L-glutamate. |
- General function:
- Involved in hydrolase activity
- Specific function:
- Degradation of external UDP-glucose to uridine monophosphate and glucose-1-phosphate, which can then be used by the cell
- Gene Name:
- ushA
- Uniprot ID:
- P07024
- Molecular weight:
- 60824
Reactions
| UDP-sugar + H(2)O = UMP + alpha-D-aldose 1-phosphate. |
| A 5'-ribonucleotide + H(2)O = a ribonucleoside + phosphate. |
- General function:
- Involved in hydrolase activity
- Specific function:
- Nucleotidase with a broad substrate specificity as it can dephosphorylate various ribo- and deoxyribonucleoside 5'- monophosphates and ribonucleoside 3'-monophosphates with highest affinity to 3'-AMP. Also hydrolyzes polyphosphate (exopolyphosphatase activity) with the preference for short-chain- length substrates (P20-25). Might be involved in the regulation of dNTP and NTP pools, and in the turnover of 3'-mononucleotides produced by numerous intracellular RNases (T1, T2, and F) during the degradation of various RNAs. Also plays a significant physiological role in stress-response and is required for the survival of E.coli in stationary growth phase
- Gene Name:
- surE
- Uniprot ID:
- P0A840
- Molecular weight:
- 26900
Reactions
| A 5'-ribonucleotide + H(2)O = a ribonucleoside + phosphate. |
| A 3'-ribonucleotide + H(2)O = a ribonucleoside + phosphate. |
| (Polyphosphate)(n) + H(2)O = (polyphosphate)(n-1) + phosphate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Nucleotidase that shows high phosphatase activity toward three nucleoside 5'-monophosphates, UMP, dUMP, and dTMP, and very low activity against TDP, IMP, UDP, GMP, dGMP, AMP, dAMP, and 6- phosphogluconate. Is strictly specific to substrates with 5'- phosphates and shows no activity against nucleoside 2'- or 3'- monophosphates. Might be involved in the pyrimidine nucleotide substrate cycles
- Gene Name:
- yjjG
- Uniprot ID:
- P0A8Y1
- Molecular weight:
- 25300
Reactions
| A 5'-ribonucleotide + H(2)O = a ribonucleoside + phosphate. |
- General function:
- Involved in hypoxanthine phosphoribosyltransferase activity
- Specific function:
- This enzyme acts exclusively on hypoxanthine; it does not act on guanine
- Gene Name:
- hpt
- Uniprot ID:
- P0A9M2
- Molecular weight:
- 20115
Reactions
| IMP + diphosphate = hypoxanthine + 5-phospho-alpha-D-ribose 1-diphosphate. |
- General function:
- Involved in nucleoside metabolic process
- Specific function:
- Acts on guanine, xanthine and to a lesser extent hypoxanthine
- Gene Name:
- gpt
- Uniprot ID:
- P0A9M5
- Molecular weight:
- 16971
Reactions
| XMP + diphosphate = 5-phospho-alpha-D-ribose 1-diphosphate + xanthine. |
- General function:
- Involved in catalytic activity
- Specific function:
- Inosine 5'-phosphate + NAD(+) + H(2)O = xanthosine 5'-phosphate + NADH
- Gene Name:
- guaB
- Uniprot ID:
- P0ADG7
- Molecular weight:
- 52022
Reactions
| Inosine 5'-phosphate + NAD(+) + H(2)O = xanthosine 5'-phosphate + NADH. |
- General function:
- Involved in acid phosphatase activity
- Specific function:
- Dephosphorylates several organic phosphomonoesters and catalyzes the transfer of low-energy phosphate groups from phosphomonoesters to hydroxyl groups of various organic compounds. Preferentially acts on aryl phosphoesters. Might function as a broad-spectrum dephosphorylating enzyme able to scavenge both 3'- and 5'-nucleotides and also additional organic phosphomonoesters
- Gene Name:
- aphA
- Uniprot ID:
- P0AE22
- Molecular weight:
- 26103
Reactions
| A phosphate monoester + H(2)O = an alcohol + phosphate. |
- General function:
- Involved in hydrolase activity
- Specific function:
- Hydrolyzes O6 atom-containing purine bases deoxyinosine triphosphate (dITP) and xanthosine triphosphate (XTP) as well as 2'-deoxy-N-6-hydroxylaminopurine triposphate (dHAPTP) to nucleotide monophosphate and pyrophosphate. Probably excludes non- standard purines from DNA precursor pool, preventing thus incorporation into DNA and avoiding chromosomal lesions
- Gene Name:
- rdgB
- Uniprot ID:
- P52061
- Molecular weight:
- 21039
Reactions
| A nucleoside triphosphate + H(2)O = a nucleotide + diphosphate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Nucleotidase that shows strict specificity toward deoxyribonucleoside 5'-monophosphates and does not dephosphorylate 5'-ribonucleotides or ribonucleoside 3'-monophosphates. Might be involved in the regulation of all dNTP pools in E.coli
- Gene Name:
- yfbR
- Uniprot ID:
- P76491
- Molecular weight:
- 22708
Reactions
| A 5'-ribonucleotide + H(2)O = a ribonucleoside + phosphate. |
Transporters
- General function:
- Involved in transporter activity
- Specific function:
- Non-specific porin
- Gene Name:
- ompN
- Uniprot ID:
- P77747
- Molecular weight:
- 41220
- General function:
- Involved in transporter activity
- Specific function:
- Uptake of inorganic phosphate, phosphorylated compounds, and some other negatively charged solutes
- Gene Name:
- phoE
- Uniprot ID:
- P02932
- Molecular weight:
- 38922
- General function:
- Involved in transporter activity
- Specific function:
- OmpF is a porin that forms passive diffusion pores which allow small molecular weight hydrophilic materials across the outer membrane. It is also a receptor for the bacteriophage T2
- Gene Name:
- ompF
- Uniprot ID:
- P02931
- Molecular weight:
- 39333
- General function:
- Involved in transporter activity
- Specific function:
- Forms passive diffusion pores which allow small molecular weight hydrophilic materials across the outer membrane
- Gene Name:
- ompC
- Uniprot ID:
- P06996
- Molecular weight:
- 40368