| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 13:53:46 -0600 |
|---|
| Update Date | 2015-10-23 17:32:16 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
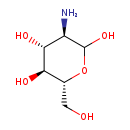
| Name: | Glucosamine |
|---|
| Description | Glucosamine (C6H14NO5) is an amino sugar that is an important precursor in the biochemical synthesis of glycosylated proteins and lipids. Glucosamine is one of the most abundant monosaccharides. (Wikipedia) |
|---|
| Structure | |
|---|
| Synonyms: | - 2-Amino-2-deoxy-D-glucopyranose
- 2-Amino-2-deoxy-D-Glucose
- 2-Amino-2-deoxyglucose
- 2-Aminoglucose
- 2-Deoxy-2-amino-D-glucose
- 2-Deoxy-2-aminoglucose
- Chitosamine
- Cosamin
- D-(+)-Glucosamine
- D-Glucosamine
- Glucosamine
|
|---|
| Chemical Formula: | C6H13NO5 |
|---|
| Weight: | Average: 179.1711
Monoisotopic: 179.079372531 |
|---|
| InChI Key: | MSWZFWKMSRAUBD-IVMDWMLBSA-N |
|---|
| InChI: | InChI=1S/C6H13NO5/c7-3-5(10)4(9)2(1-8)12-6(3)11/h2-6,8-11H,1,7H2/t2-,3-,4-,5-,6?/m1/s1 |
|---|
| CAS number: | 3416-24-8 |
|---|
| IUPAC Name: | (3R,4R,5S,6R)-3-amino-6-(hydroxymethyl)oxane-2,4,5-triol |
|---|
| Traditional IUPAC Name: | glucosamine |
|---|
| SMILES: | N[C@H]1C(O)O[C@H](CO)[C@@H](O)[C@@H]1O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as hexoses. These are monosaccharides in which the sugar unit is a is a six-carbon containing moeity. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Hexoses |
|---|
| Alternative Parents | |
|---|
| Substituents | - Hexose monosaccharide
- Amino saccharide
- Oxane
- 1,2-aminoalcohol
- Hemiacetal
- Secondary alcohol
- Polyol
- Organoheterocyclic compound
- Oxacycle
- Primary amine
- Primary alcohol
- Organopnictogen compound
- Organonitrogen compound
- Primary aliphatic amine
- Organic nitrogen compound
- Alcohol
- Amine
- Hydrocarbon derivative
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | 1 |
|---|
| Melting point: | 88 °C |
|---|
| Experimental Properties: | | Property | Value | Source |
|---|
| Water Solubility: | 330 mg/mL [HMP experimental] | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | - Amino sugar and nucleotide sugar metabolism ec00520
- Phosphotransferase system (PTS) ec02060
|
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| | Concentration | Strain | Media | Growth Status | Growth System | Temperature | Details |
|---|
| 19± 0 uM | BW25113 | 48 mM Na2HPO4, 22 mM KH2PO4, 10 mM NaCl, 45 mM (NH4)2SO4, supplemented with 1 mM MgSO4, 1 mg/l thiamine·HCl, 5.6 mg/l CaCl2, 8 mg/l FeCl3, 1 mg/l MnCl2·4H2O, 1.7 mg/l ZnCl2, 0.43 mg/l CuCl2·2H2O, 0.6 mg/l CoCl2·2H2O and 0.6 mg/l Na2MoO4·2H2O. 4 g/L Gluco | Stationary Phase, glucose limited | Bioreactor, pH controlled, O2 and CO2 controlled, dilution rate: 0.2/h | 37 oC | PMID: 17379776 |
|
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-06tr-9300000000-7e2a89f48724cdda814c | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-0ul9-9847700000-b4779809586433e7ec78 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-03di-4900000000-364d52fc6e7e423f52bb | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-00di-9000000000-762e016c89a40a9e3f49 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-00di-9000000000-bd04d561d78f2abad229 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-03di-9500000000-9bfc2897a4b66f71c7a1 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-0007-9000000000-f842b779439be8970cd9 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-074i-9000000000-622ec0743231c0cbaca6 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-03e9-9700000000-5087af92679b2640ba6c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-b9ba212915cbdaed8628 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-2d1825b518f09745e036 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-03la-9100000000-3da7ee17d7512a59e3ef | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-03e9-9400000000-cca17ef05a47a7be4d43 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-01x0-9500000000-5fbd02d318d3ec58f98b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 30V, Positive | splash10-0abc-9000000000-11e86d324c183174364e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 0V, Positive | splash10-001i-0900000000-3ba18bb57823024db64e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-03e9-9500000000-52f44695a6cfb6459105 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-03di-9800000000-264ee54a99e01e0d4e30 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-00dl-9000000000-08f87dc3e44979542ca2 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-06r6-9000000000-635fa893be82ffdeca97 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-03e9-9600000000-09232c8766303805f765 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01q9-0900000000-4c88765919efa0ec2d2d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03ea-6900000000-301590fbe90692448d5e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052g-9100000000-d6a51790ce1df14a067a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-002r-9400000000-e43a7a5cb44f6c461544 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-053r-9800000000-56b95ef43b164db5524d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052f-9000000000-1434c5ce1e9649c1cbf1 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H,1H] 2D NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Ishii, N., Nakahigashi, K., Baba, T., Robert, M., Soga, T., Kanai, A., Hirasawa, T., Naba, M., Hirai, K., Hoque, A., Ho, P. Y., Kakazu, Y., Sugawara, K., Igarashi, S., Harada, S., Masuda, T., Sugiyama, N., Togashi, T., Hasegawa, M., Takai, Y., Yugi, K., Arakawa, K., Iwata, N., Toya, Y., Nakayama, Y., Nishioka, T., Shimizu, K., Mori, H., Tomita, M. (2007). "Multiple high-throughput analyses monitor the response of E. coli to perturbations." Science 316:593-597. Pubmed: 17379776
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
- Winder, C. L., Dunn, W. B., Schuler, S., Broadhurst, D., Jarvis, R., Stephens, G. M., Goodacre, R. (2008). "Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites." Anal Chem 80:2939-2948. Pubmed: 18331064
|
|---|
| Synthesis Reference: | Li, Nan; Li, Jiheng. Preparation of D-glucosamine hydrochloride. Zhongguo Yaoke Daxue Xuebao (1997), 28(1), 56-58. |
|---|
| Material Safety Data Sheet (MSDS) | Download (PDF) |
|---|
| Links |
|---|
| External Links: | |
|---|