| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 13:47:43 -0600 |
|---|
| Update Date | 2015-09-13 12:56:10 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
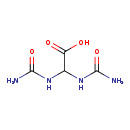
| Name: | Allantoic acid |
|---|
| Description | Allantoic acid is the END product of Allantoicase [EC:3.5.3.4], an enzyme involved in uric acid degradation (Purine metabolism). It is a crystalline acid obtained by hydrolysis of allantoin. (Wikipedia) |
|---|
| Structure | |
|---|
| Synonyms: | - Allantoate
- Allantoic acid
- Diureidoacetate
- Diureidoacetic acid
|
|---|
| Chemical Formula: | C4H8N4O4 |
|---|
| Weight: | Average: 176.1307
Monoisotopic: 176.054554764 |
|---|
| InChI Key: | NUCLJNSWZCHRKL-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/C4H8N4O4/c5-3(11)7-1(2(9)10)8-4(6)12/h1H,(H,9,10)(H3,5,7,11)(H3,6,8,12) |
|---|
| CAS number: | 99-16-1 |
|---|
| IUPAC Name: | 2,2-bis(carbamoylamino)acetic acid |
|---|
| Traditional IUPAC Name: | allantoic acid |
|---|
| SMILES: | NC(=O)NC(NC(N)=O)C(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as n-carbamoyl-alpha amino acids. N-carbamoyl-alpha amino acids are compounds containing an alpha amino acid which bears an carbamoyl group at its terminal nitrogen atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | N-carbamoyl-alpha amino acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - N-carbamoyl-alpha-amino acid
- Urea
- Carbonic acid derivative
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Organic nitrogen compound
- Organic oxygen compound
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -1 |
|---|
| Melting point: | 180-181 °C |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | | allantoin degradation (anaerobic) | PW002050 | 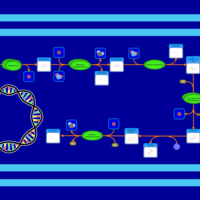 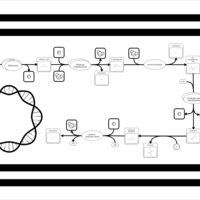 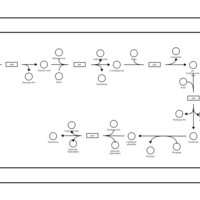 | | glycolate and glyoxylate degradation | PW000827 | 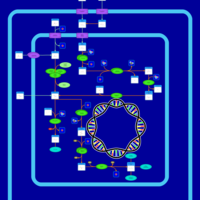 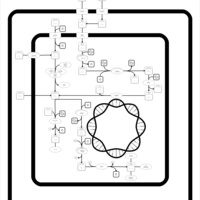 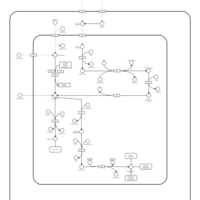 |
|
|---|
| KEGG Pathways: | |
|---|
| EcoCyc Pathways: | - allantoin degradation to ureidoglycolate II (ammonia producing) PWY-5698
|
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (Non-derivatized) | splash10-0002-1900000000-21581f2d921374c16317 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0uej-0911000000-7b31dde6fb9c6e9ac110 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0f7a-0900000000-d8f1371697a09dc2d063 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0udi-0920000000-7557e41727738c553aef | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-000x-9800000000-0da9967a35111f5e77c5 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-006x-9320000000-3e52483917d7449c6f21 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-03di-9500000000-311fe8e2669139e60084 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-03k9-9000000000-eb594a4f54a39f5aaad4 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-03di-9000000000-24cb204bd4d824cf2b53 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-003r-0900000000-91ad7fef4b504549f849 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-000i-9500000000-1f86fa2bb6e6a3e1988e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-000i-9000000000-6f90d802bf75ec4bc187 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-00dr-9000000000-3900e78a2acf61f39eff | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-00dl-9000000000-e98594c1e1b2aa50540a | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-003r-0900000000-91ad7fef4b504549f849 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-000i-9500000000-1f86fa2bb6e6a3e1988e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-000i-9000000000-6f90d802bf75ec4bc187 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-00dr-9000000000-3900e78a2acf61f39eff | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-00dl-9000000000-e98594c1e1b2aa50540a | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-03dl-9000000000-c2a7cae004d98357168a | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 30V, Positive | splash10-03di-9000000000-31fc32546236ce67c00c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-000i-9000000000-27a2a739677caaa22b0a | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-000i-9400000000-120384794acaedafde99 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-00dl-9000000000-bf94440d44163d36a2e4 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-004r-9500000000-f32dafebab7d351734b5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-003r-1900000000-e1f2f46a56a37aa06912 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00li-9600000000-ee4b95c0d08c896e3843 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-9300000000-1beb69c57c7dfcea4351 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001r-9800000000-682ec8d4340d0075c369 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a5i-9400000000-6faaacd64e9faecfb1bf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9100000000-ac2cde105e2954f49b46 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
- Vigetti D, Pollegioni L, Monetti C, Prati M, Bernardini G, Gornati R: Property comparison of recombinant amphibian and mammalian allantoicases. FEBS Lett. 2002 Feb 13;512(1-3):323-8. Pubmed: 11852104
- Winder, C. L., Dunn, W. B., Schuler, S., Broadhurst, D., Jarvis, R., Stephens, G. M., Goodacre, R. (2008). "Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites." Anal Chem 80:2939-2948. Pubmed: 18331064
|
|---|
| Synthesis Reference: | Hermanowicz, Witold. Allantoic acid. Formation of allantoic acid from allantoin. Roczniki Chemii (1948), 22 159-80. |
|---|
| Material Safety Data Sheet (MSDS) | Download (PDF) |
|---|
| Links |
|---|
| External Links: | |
|---|