Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
Hydrocinnamic acid (M2MDB000184)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-05-31 13:01:05 -0600 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-09-13 12:56:09 -0600 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Hydrocinnamic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Hydrocinnamic acid is an analogue of phenylalanine. It is a substrate of the enzyme oxidoreductases [EC 1.14.12.-] in the pathway phenylalanine metabolism (KEGG). | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
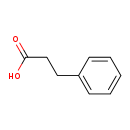
| Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C9H10O2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 150.1745 Monoisotopic: 150.068079564 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | XMIIGOLPHOKFCH-UHFFFAOYSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C9H10O2/c10-9(11)7-6-8-4-2-1-3-5-8/h1-5H,6-7H2,(H,10,11) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 501-52-0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | 3-phenylpropanoic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | 3-phenylpropionic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | OC(=O)CCC1=CC=CC=C1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as phenylpropanoic acids. Phenylpropanoic acids are compounds with a structure containing a benzene ring conjugated to a propanoic acid. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Phenylpropanoids and polyketides | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Phenylpropanoic acids | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Phenylpropanoic acids | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic homomonocyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 45-48 °C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Hydrogen ion + NADH + Oxygen + Hydrocinnamic acid > Cis-3-(3-carboxyethyl)-3,5-cyclohexadiene-1,2-diol + NAD Hydrocinnamic acid + Oxygen + NADH + Hydrogen ion <> cis-3-(Carboxy-ethyl)-3,5-cyclo-hexadiene-1,2-diol + NAD Hydrocinnamic acid + NADH + Oxygen + Hydrogen ion > cis-3-(Carboxy-ethyl)-3,5-cyclo-hexadiene-1,2-diol + NAD Hydrocinnamic acid + NADH + Oxygen > cis-3-(Carboxy-ethyl)-3,5-cyclo-hexadiene-1,2-diol + NAD NADH + Hydrogen ion + Oxygen + Hydrocinnamic acid <> cis-3-(Carboxy-ethyl)-3,5-cyclo-hexadiene-1,2-diol + NAD + Trans-2,3-Dihydroxycinnamate Hydrocinnamic acid + NADH + Oxygen <> NAD + cis-3-(Carboxy-ethyl)-3,5-cyclo-hexadiene-1,2-diol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Muller, August John; Bowers, Joseph Stanton, Jr.; Eubanks, John Robert Ira; Geiger, Carey Cecil; Santobianco, John Gabriel. Process for preparing hydrocinnamic acid from cinnamaldehyde. PCT Int. Appl. (1999), 18 pp. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in iron ion binding
- Specific function:
- Part of the multicomponent 3-phenylpropionate dioxygenase. Converts 3-phenylpropionic acid (PP) and cinnamic acid (CI) into 3-phenylpropionate-dihydrodiol (PP-dihydrodiol) and cinnamic acid-dihydrodiol (CI-dihydrodiol), respectively
- Gene Name:
- hcaE
- Uniprot ID:
- P0ABR5
- Molecular weight:
- 51109
Reactions
| 3-phenylpropanoate + NADH + O(2) = 3-(cis-5,6-dihydroxycyclohexa-1,3-dien-1-yl)propanoate + NAD(+). |
| (2E)-3-phenylprop-2-enoate + NADH + O(2) = (2E)-3-(2,3-dihydroxyphenyl)prop-2-enoate + NAD(+). |
- General function:
- Involved in oxidoreductase activity
- Specific function:
- Part of the multicomponent 3-phenylpropionate dioxygenase, that converts 3-phenylpropionic acid (PP) and cinnamic acid (CI) into 3-phenylpropionate-dihydrodiol (PP- dihydrodiol) and cinnamic acid-dihydrodiol (CI-dihydrodiol), respectively. This protein seems to be a 2Fe-2S ferredoxin
- Gene Name:
- hcaC
- Uniprot ID:
- P0ABW0
- Molecular weight:
- 11329
- General function:
- Involved in oxidoreductase activity
- Specific function:
- Part of the multicomponent 3-phenylpropionate dioxygenase, that converts 3-phenylpropionic acid (PP) and cinnamic acid (CI) into 3-phenylpropionate-dihydrodiol (PP- dihydrodiol) and cinnamic acid-dihydrodiol (CI-dihydrodiol), respectively
- Gene Name:
- hcaD
- Uniprot ID:
- P77650
- Molecular weight:
- 43978
Reactions
| Reduced ferredoxin + NAD(+) = oxidized ferredoxin + NADH. |
- General function:
- Involved in 3-phenylpropionate dioxygenase activity
- Specific function:
- Part of the multicomponent 3-phenylpropionate dioxygenase. Converts 3-phenylpropionic acid (PP) and cinnamic acid (CI) into 3-phenylpropionate-dihydrodiol (PP-dihydrodiol) and cinnamic acid-dihydrodiol (CI-dihydrodiol), respectively
- Gene Name:
- hcaF
- Uniprot ID:
- Q47140
- Molecular weight:
- 20579
Reactions
| 3-phenylpropanoate + NADH + O(2) = 3-(cis-5,6-dihydroxycyclohexa-1,3-dien-1-yl)propanoate + NAD(+). |
| (2E)-3-phenylprop-2-enoate + NADH + O(2) = (2E)-3-(2,3-dihydroxyphenyl)prop-2-enoate + NAD(+). |
Transporters
- General function:
- Involved in transmembrane transport
- Specific function:
- Probable permease involved in the uptake of 3- phenylpropionic acid
- Gene Name:
- hcaT
- Uniprot ID:
- Q47142
- Molecular weight:
- 41592
- General function:
- Involved in transmembrane transport
- Specific function:
- Could be a transporter for 3-phenylpropionate (hydrocinnamic acid)
- Gene Name:
- mhpT
- Uniprot ID:
- P77589
- Molecular weight:
- 41550
- General function:
- Involved in transporter activity
- Specific function:
- Non-specific porin
- Gene Name:
- ompN
- Uniprot ID:
- P77747
- Molecular weight:
- 41220
- General function:
- Involved in transporter activity
- Specific function:
- Uptake of inorganic phosphate, phosphorylated compounds, and some other negatively charged solutes
- Gene Name:
- phoE
- Uniprot ID:
- P02932
- Molecular weight:
- 38922
- General function:
- Involved in transporter activity
- Specific function:
- OmpF is a porin that forms passive diffusion pores which allow small molecular weight hydrophilic materials across the outer membrane. It is also a receptor for the bacteriophage T2
- Gene Name:
- ompF
- Uniprot ID:
- P02931
- Molecular weight:
- 39333
- General function:
- Involved in transporter activity
- Specific function:
- Forms passive diffusion pores which allow small molecular weight hydrophilic materials across the outer membrane
- Gene Name:
- ompC
- Uniprot ID:
- P06996
- Molecular weight:
- 40368