Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
D-Xylulose (M2MDB000165)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-05-31 10:28:15 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-09-13 12:56:08 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
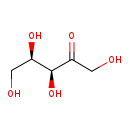
| Name: | D-Xylulose | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | D-xylulose is a monosaccharide containing five carbon atoms. D-xylulose is converted from xylitol by the enzyme NAD+-linked xylitol dehydrogenase (EC 1.1.1.9) in the glucuronate pathway. Most likely, D-xylulose (as well as D-arabinose or D-ribulose) is a precursor of the pentiol D-arabitol, since pentitols are derived from their corresponding pentose phosphate precursors via pentoses. Although pentitols are present in all living organisms, knowledge concerning their metabolism is limited. (PMID: 15234337, Mol Genet Metab. 2004 Jul;82(3):231-7.) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C5H10O5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 150.1299 Monoisotopic: 150.05282343 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | ZAQJHHRNXZUBTE-WUJLRWPWSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C5H10O5/c6-1-3(8)5(10)4(9)2-7/h3,5-8,10H,1-2H2/t3-,5+/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 551-84-8 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (3S,4R)-1,3,4,5-tetrahydroxypentan-2-one | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | D-xylulose | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | [H][C@@](O)(CO)[C@]([H])(O)C(=O)CO | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as pentoses. These are monosaccharides in which the carbohydrate moiety contains five carbon atoms. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic oxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Carbohydrates and carbohydrate conjugates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Pentoses | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Liquid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 15 °C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosine triphosphate + D-Xylulose <> ADP + Hydrogen ion + Xylulose 5-phosphate D-Xylose <> D-Xylulose Adenosine triphosphate + D-Xylulose <> ADP + Xylulose 5-phosphate D-xylose <> D-Xylulose D-Xylulose + Adenosine triphosphate > Hydrogen ion + Xylulose 5-phosphate + ADP D-Xylose > D-Xylulose Adenosine triphosphate + D-Xylulose > ADP + Xylulose 5-phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Takeuchi, Sonoko; Tonouchi, Naoto; Yokozeki, Kenzo. Production of xylitol or D-xylulose by fermentation of glucose with Gluconobacter, Acetobacter, and Frateuria. Eur. Pat. Appl. (2000), 10 pp. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in xylose isomerase activity
- Specific function:
- D-xylose = D-xylulose
- Gene Name:
- xylA
- Uniprot ID:
- P00944
- Molecular weight:
- 49742
Reactions
| D-xylose = D-xylulose. |
- General function:
- Involved in phosphotransferase activity, alcohol group as acceptor
- Specific function:
- ATP + L(or D)-ribulose = ADP + L(or D)- ribulose 5-phosphate
- Gene Name:
- araB
- Uniprot ID:
- P08204
- Molecular weight:
- 61089
Reactions
| ATP + L(or D)-ribulose = ADP + L(or D)-ribulose 5-phosphate. |
- General function:
- Involved in phosphotransferase activity, alcohol group as acceptor
- Specific function:
- ATP + D-xylulose = ADP + D-xylulose 5- phosphate
- Gene Name:
- xylB
- Uniprot ID:
- P09099
- Molecular weight:
- 52618
Reactions
| ATP + D-xylulose = ADP + D-xylulose 5-phosphate. |
Transporters
- General function:
- Involved in transmembrane transport
- Specific function:
- Involved in the efflux of sugars. The physiological role may be the detoxification of non-metabolizable sugar analogs
- Gene Name:
- setC
- Uniprot ID:
- P31436
- Molecular weight:
- 43493