| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 10:28:05 -0600 |
|---|
| Update Date | 2015-09-13 12:56:08 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | Galactaric acid |
|---|
| Description | Galactaric acid is the dicarboxylic sugar acid resulting from oxidation glactose with dilute nitric acid. It is a substrate of galactarate O-hydroxycinnamoyltransferase [EC 2.3.1.130]. (KEGG) |
|---|
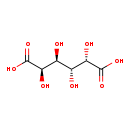
| Structure | |
|---|
| Synonyms: | - (2R,3S,4R,5S)-2,3,4,5-tetrahydroxyhexanedioate
- (2R,3S,4R,5S)-2,3,4,5-tetrahydroxyhexanedioic acid
- meso-galactarate
- D-Galactarate
- D-Galactaric acid
- Galactarate
- Galactaric acid
- Galactosaccharate
- Galactosaccharic acid
- Hexarate
- Hexaric acid
- Meso-galactarate
- Meso-galactaric acid
- Mucate
- Mucic acid
- Saccharolactate
- Saccharolactic acid
- Schleimsaure
|
|---|
| Chemical Formula: | C6H10O8 |
|---|
| Weight: | Average: 210.1388
Monoisotopic: 210.037567296 |
|---|
| InChI Key: | DSLZVSRJTYRBFB-DUHBMQHGSA-N |
|---|
| InChI: | InChI=1S/C6H10O8/c7-1(3(9)5(11)12)2(8)4(10)6(13)14/h1-4,7-10H,(H,11,12)(H,13,14)/t1-,2+,3+,4- |
|---|
| CAS number: | 526-99-8 |
|---|
| IUPAC Name: | (2R,3S,4R,5S)-2,3,4,5-tetrahydroxyhexanedioic acid |
|---|
| Traditional IUPAC Name: | mucin |
|---|
| SMILES: | O[C@@H]([C@@H](O)[C@H](O)C(O)=O)[C@@H](O)C(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glucuronic acid derivatives. Glucuronic acid derivatives are compounds containing a glucuronic acid moiety (or a derivative), which consists of a glucose moiety with the C6 carbon oxidized to a carboxylic acid. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Glucuronic acid derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glucuronic acid or derivatives
- Medium-chain hydroxy acid
- Medium-chain fatty acid
- Beta-hydroxy acid
- Hydroxy fatty acid
- Monosaccharide
- Hydroxy acid
- Dicarboxylic acid or derivatives
- Alpha-hydroxy acid
- Fatty acyl
- Fatty acid
- Secondary alcohol
- Polyol
- Carboxylic acid
- Carboxylic acid derivative
- Hydrocarbon derivative
- Organic oxide
- Carbonyl group
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -2 |
|---|
| Melting point: | 220-225 °C |
|---|
| Experimental Properties: | | Property | Value | Source |
|---|
| Water Solubility: | 3.3 mg/mL at 14 oC [YALKOWSKY,SH & DANNENFELSER,RM (1992)] | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | | superpathway of D-glucarate and D-galactarate degradation | PW000795 |  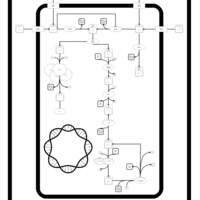 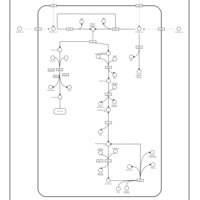 |
|
|---|
| KEGG Pathways: | - Ascorbate and aldarate metabolism ec00053
|
|---|
| EcoCyc Pathways: | |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (6 TMS) | splash10-001l-0987000000-76d3c6748cb8a79adc39 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-001l-0987000000-76d3c6748cb8a79adc39 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-05bf-9400000000-b80ffbca6c943091f16e | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (6 TMS) - 70eV, Positive | splash10-00b9-6011394000-83b916b3001ad13cf34d | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_4_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_5_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_6_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("Galactaric acid,4TBDMS,#8" TMS) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Negative (Annotated) | splash10-052r-9260000000-16fac7d5f0f85285824d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Negative (Annotated) | splash10-0abi-9000000000-80a4d5a9910d243f2e48 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Negative (Annotated) | splash10-0abc-9000000000-144a5e96a20e3c66ca9d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-0a4i-0190000000-53cb6532b098f7615896 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-052r-9430000000-7bc8b56c9c00f7aed59f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-000i-9000000000-34bca663662f41b51683 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-05fr-9000000000-c1a4fdc6f3d7d8e237aa | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-05fr-9000000000-f2f36dc0c7eca0f78710 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-052r-9240000000-c24ca4338feeca20c7f5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0a4i-0190000000-53cb6532b098f7615896 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-052r-9430000000-7bc8b56c9c00f7aed59f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-000i-9000000000-34bca663662f41b51683 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-05fr-9000000000-c1a4fdc6f3d7d8e237aa | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-05fr-9000000000-f2f36dc0c7eca0f78710 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-052r-9240000000-c24ca4338feeca20c7f5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-000i-9210000000-2f7e7ffe11298f785adc | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-052r-9330000000-afb6ef48335ee358a6c7 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-05fu-9000000000-fe6896a18ce47504bbf5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-052r-9000000000-6c2579c066e15d190bad | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03dl-1940000000-66a37b8eabfbc367df77 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4r-9700000000-ae255ce964e55a9644c5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a6r-9100000000-fa70d3c1751580028d65 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0adr-9820000000-19f2dbe53ca43b0a1195 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4r-9500000000-e0ce27927a6119cf1489 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0ap0-9100000000-0c338926a15f642737e1 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
- Winder, C. L., Dunn, W. B., Schuler, S., Broadhurst, D., Jarvis, R., Stephens, G. M., Goodacre, R. (2008). "Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites." Anal Chem 80:2939-2948. Pubmed: 18331064
|
|---|
| Synthesis Reference: | Fauvarque, Jean-Francois; Guerin, Christophe; Petit, Serge; De Regis, Baynast. Process and electrolytic cell for preparing galactaric acid. Fr. Demande (1994), 14 pp. |
|---|
| Material Safety Data Sheet (MSDS) | Download (PDF) |
|---|
| Links |
|---|
| External Links: | |
|---|