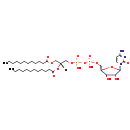| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2015-09-08 19:54:15 -0600 |
|---|
| Update Date | 2015-12-09 12:12:59 -0700 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | CDP-DG(12:0/12:0) |
|---|
| Description | CDP-DG(12:0/12:0) belongs to the family of CDP-diacylglycerols. It is a glycerophospholipid containing a diacylglycerol, with a cytidine diphosphate attached to the oxygen O1 or O2 of the glycerol part. As is the case with diacylglycerols, phosphatidylserines can have many different combinations of fatty acids of varying lengths and saturation attached to the C-1 and C-2 positions. CDP-DG(12:0/12:0), in particular, consists of two dodecanoyl chain at positions C-1 and C2. In E. coli glycerophospholipid metabolism, The biosynthesis of CDP-diacylglycerol (CDP-DG) involves condensation of phosphatidic acid (PA) and cytidine triphosphate, with elimination of pyrophosphate, catalysed by the enzyme CDP-diacylglycerol synthase. The resulting CDP-diacylglycerol can be utilized immediately for the synthesis of phosphatidylglycerol (PG), and thence cardiolipin (CL), and of phosphatidylinositol (PI). CDP-DG(12:0/12:0) is also a substrate of CDP-diacylglycerol pyrophosphatase. It is involved in CDP-diacylglycerol degradation pathway. |
|---|
| Structure | |
|---|
| Synonyms: | Not Available |
|---|
| Chemical Formula: | C36H65N3O15P2 |
|---|
| Weight: | Average: 841.87
Monoisotopic: 841.389092401 |
|---|
| InChI Key: | PTPPKXVNJJIECF-MYNNLVAUSA-N |
|---|
| InChI: | InChI=1S/C36H65N3O15P2/c1-3-5-7-9-11-13-15-17-19-21-31(40)49-25-28(52-32(41)22-20-18-16-14-12-10-8-6-4-2)26-50-55(45,46)54-56(47,48)51-27-29-33(42)34(43)35(53-29)39-24-23-30(37)38-36(39)44/h23-24,28-29,33-35,42-43H,3-22,25-27H2,1-2H3,(H,45,46)(H,47,48)(H2,37,38,44)/t28-,29-,33+,34?,35-/m1/s1 |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | {[(2R,3R,5R)-5-(4-amino-2-oxo-1,2-dihydropyrimidin-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}({[(2R)-2,3-bis(dodecanoyloxy)propoxy](hydroxy)phosphoryl}oxy)phosphinic acid |
|---|
| Traditional IUPAC Name: | [(2R,3R,5R)-5-(4-amino-2-oxopyrimidin-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy([(2R)-2,3-bis(dodecanoyloxy)propoxy(hydroxy)phosphoryl]oxy)phosphinic acid |
|---|
| SMILES: | [H][C@@](COC(=O)CCCCCCCCCCC)(COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H](C(O)[C@H]1O)N1C=CC(N)=NC1=O)OC(=O)CCCCCCCCCCC |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as cdp-diacylglycerols. These are glycerolipids containing a diacylglycerol, with a cytidine diphosphate attached to the oxygen O1 or O2 of the glycerol part. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerophospholipids |
|---|
| Sub Class | CDP-glycerols |
|---|
| Direct Parent | CDP-diacylglycerols |
|---|
| Alternative Parents | |
|---|
| Substituents | - Cdp-diacylglycerol
- Pyrimidine ribonucleoside diphosphate
- Diacyl-glycerol-3-pyrophosphate
- Pentose phosphate
- Pentose-5-phosphate
- Glycosyl compound
- N-glycosyl compound
- Monosaccharide phosphate
- Organic pyrophosphate
- Pentose monosaccharide
- Aminopyrimidine
- Fatty acid ester
- Pyrimidone
- Monoalkyl phosphate
- Imidolactam
- Pyrimidine
- Fatty acyl
- Alkyl phosphate
- Phosphoric acid ester
- Dicarboxylic acid or derivatives
- Hydropyrimidine
- Organic phosphoric acid derivative
- Monosaccharide
- Tetrahydrofuran
- Heteroaromatic compound
- Secondary alcohol
- Amino acid or derivatives
- 1,2-diol
- Carboxylic acid ester
- Organoheterocyclic compound
- Azacycle
- Oxacycle
- Carboxylic acid derivative
- Alcohol
- Organonitrogen compound
- Organic nitrogen compound
- Organooxygen compound
- Amine
- Carbonyl group
- Primary amine
- Organic oxygen compound
- Organopnictogen compound
- Hydrocarbon derivative
- Organic oxide
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | -2 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Membrane |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | |
|---|
| References |
|---|
| References: | - Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- Uniprot Consortium (2012). "Reorganizing the protein space at the Universal Protein Resource (UniProt)." Nucleic Acids Res 40:D71-D75. Pubmed: 22102590
- Yurtsever D. (2007). Fatty acid methyl ester profiling of Enterococcus and Esherichia coli for microbial source tracking. M.sc. Thesis. Villanova University: U.S.A
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | | Resource | Link |
|---|
| CHEBI ID | Not Available | | HMDB ID | Not Available | | Pubchem Compound ID | 3247021 | | Kegg ID | Not Available | | ChemSpider ID | Not Available | | Wikipedia ID | Not Available | | BioCyc ID | Not Available |
|
|---|