| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2015-09-08 19:03:02 -0600 |
|---|
| Update Date | 2015-09-16 17:35:11 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
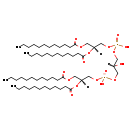
| Name: | CL(12:0/12:0/12:0/12:0) |
|---|
| Description | CL(12:0/12:0/12:0/12:0) is a cardiolipin (CL). Cardiolipins are sometimes called a 'double' phospholipid because they have four fatty acid tails, instead of the usual two. CL(12:0/12:0/12:0/12:0) contains four chains of dodecanoic acid at the C1, C2, C3 and C4 positions. While the theoretical charge of cardiolipins is -2, under normal physiological conditions (pH near 7), the molecule may carry only one negative charge. In prokaryotes such as E. coli, the enzyme known as diphosphatidylglycerol synthase catalyses the transfer of the phosphatidyl moiety of one phosphatidylglycerol to the free 3'-hydroxyl group of another, with the elimination of one molecule of glycerol. In E. coli, which acylates its glycerophospholipids with acyl chains ranging in length from 12 to 19 carbons and possibly containing an unsaturation, or a cyclopropane group more than 100 possible CL molecular species are theoretically possible. E. coli membranes consist of ~5% cardiolipin (CL), 20-25% phosphatidylglycerol (PG), and 70-80% phosphatidylethanolamine (PE) as well as smaller amounts of phosphatidylserine (PS). CL is distributed between the two leaflets of the bilayers and is located preferentially at the poles and septa in E. coli and other rod-shaped bacteria. It is known that the polar positioning of the proline transporter ProP and the mechanosensitive ion channel MscS in E. coli is dependent on CL. It is believed that cell shape may influence the localization of CL and the localization of certain membrane proteins. |
|---|
| Structure | |
|---|
| Synonyms: | Not Available |
|---|
| Chemical Formula: | C57H110O17P2 |
|---|
| Weight: | Average: 1129.4206
Monoisotopic: 1128.721825118 |
|---|
| InChI Key: | LNSKUBQTAKEJOP-TVALRIBFSA-N |
|---|
| InChI: | InChI=1S/C57H110O17P2/c1-5-9-13-17-21-25-29-33-37-41-54(59)67-47-52(73-56(61)43-39-35-31-27-23-19-15-11-7-3)49-71-75(63,64)69-45-51(58)46-70-76(65,66)72-50-53(74-57(62)44-40-36-32-28-24-20-16-12-8-4)48-68-55(60)42-38-34-30-26-22-18-14-10-6-2/h51-53,58H,5-50H2,1-4H3,(H,63,64)(H,65,66)/t52-,53-/m1/s1 |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | [(2R)-2,3-bis(dodecanoyloxy)propoxy][3-({[(2R)-2,3-bis(dodecanoyloxy)propoxy](hydroxy)phosphoryl}oxy)-2-hydroxypropoxy]phosphinic acid |
|---|
| Traditional IUPAC Name: | (2R)-2,3-bis(dodecanoyloxy)propoxy(3-{[(2R)-2,3-bis(dodecanoyloxy)propoxy(hydroxy)phosphoryl]oxy}-2-hydroxypropoxy)phosphinic acid |
|---|
| SMILES: | CCCCCCCCCCCC(=O)OC[C@H](COP(O)(=O)OCC(O)COP(O)(=O)OC[C@@H](COC(=O)CCCCCCCCCCC)OC(=O)CCCCCCCCCCC)OC(=O)CCCCCCCCCCC |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as cardiolipins. These are glycerophospholipids in which the O1 and O3 oxygen atoms of the central glycerol moiety are each linked to one 1,2-diacylglycerol chain. Their general formula is OC(COP(O)(=O)OC[C@@H](CO[R1])O[R2])COP(O)(=O)OC[C@@H](CO[R3])O[R4], where R1-R4 are four fatty acyl chains. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerophospholipids |
|---|
| Sub Class | Glycerophosphoglycerophosphoglycerols |
|---|
| Direct Parent | Cardiolipins |
|---|
| Alternative Parents | |
|---|
| Substituents | - Cardiolipin
- Tetracarboxylic acid or derivatives
- Fatty acid ester
- Dialkyl phosphate
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Alkyl phosphate
- Fatty acyl
- Carboxylic acid ester
- Secondary alcohol
- Carboxylic acid derivative
- Organooxygen compound
- Alcohol
- Organic oxide
- Organic oxygen compound
- Hydrocarbon derivative
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | -2 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Membrane |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | |
|---|
| References |
|---|
| References: | - De Siervo, A. J. (1969). "Alterations in the phospholipid composition of Escherichia coli B during growth at different temperatures." J Bacteriol 100:1342-1349. Pubmed: 4902814
- Garrett, T. A., O'Neill, A. C., Hopson, M. L. (2012). "Quantification of cardiolipin molecular species in Escherichia coli lipid extracts using liquid chromatography/electrospray ionization mass spectrometry." Rapid Commun Mass Spectrom 26:2267-2274. Pubmed: 22956318
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- Uniprot Consortium (2012). "Reorganizing the protein space at the Universal Protein Resource (UniProt)." Nucleic Acids Res 40:D71-D75. Pubmed: 22102590
- Yurtsever D. (2007). Fatty acid methyl ester profiling of Enterococcus and Esherichia coli for microbial source tracking. M.sc. Thesis. Villanova University: U.S.A
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | | Resource | Link |
|---|
| CHEBI ID | Not Available | | HMDB ID | HMDB0201179 | | Pubchem Compound ID | 102383108 | | Kegg ID | Not Available | | ChemSpider ID | 59695819 | | Wikipedia ID | Not Available | | BioCyc ID | Not Available |
|
|---|