| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2015-09-08 17:50:21 -0600 |
|---|
| Update Date | 2015-09-14 16:46:43 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
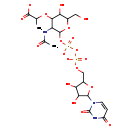
| Name: | UDP-N-acetyl-α-D-muramate |
|---|
| Description | UDP-N-acetyl-alpha-D-muramate is an intermediate in UDP-N-acetylmuramoyl-pentapeptide biosynthesis I (meso-diaminopimelate containing) pathway in E.coli. It is a substrate for the enzyme UDP-N-acetylmuramate-alanine ligase which catalyzes the reaction L-alanine + UDP-N-acetyl-alpha-D-muramate + ATP → UDP-N-acetyl-alpha-D-muramoyl-L-alanine + ADP + phosphate + H+. It is also a product for enzyme UDP-N-acetylenolpyruvoylglucosamine reductase which catalyzes reaction UDP-N-acetyl-alpha-D-glucosamine-enolpyruvate + NADPH + H+ -> UDP-N-acetyl-alpha-D-muramate + NADP+ (BioCyc compound: UDP-N-ACETYLMURAMATE). |
|---|
| Structure | |
|---|
| Synonyms: | - UDP-N-Acetyl-α-D-muramic acid
|
|---|
| Chemical Formula: | C20H28N3O19P2 |
|---|
| Weight: | Average: 676.395
Monoisotopic: 676.080870429 |
|---|
| InChI Key: | NQBRVZNDBBMBLJ-UHFFFAOYSA-K |
|---|
| InChI: | InChI=1S/C20H31N3O19P2/c1-7(18(30)31)38-16-12(21-8(2)25)19(40-9(5-24)14(16)28)41-44(35,36)42-43(33,34)37-6-10-13(27)15(29)17(39-10)23-4-3-11(26)22-20(23)32/h3-4,7,9-10,12-17,19,24,27-29H,5-6H2,1-2H3,(H,21,25)(H,30,31)(H,33,34)(H,35,36)(H,22,26,32)/p-3 |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | 2-[(2-{[({[5-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-1-yl)-3,4-dihydroxyoxolan-2-yl]methyl phosphonato}oxy)phosphinato]oxy}-3-acetamido-5-hydroxy-6-(hydroxymethyl)oxan-4-yl)oxy]propanoate |
|---|
| Traditional IUPAC Name: | 2-({2-[({[5-(2,4-dioxo-3H-pyrimidin-1-yl)-3,4-dihydroxyoxolan-2-yl]methyl phosphonato}oxyphosphinato)oxy]-3-acetamido-5-hydroxy-6-(hydroxymethyl)oxan-4-yl}oxy)propanoate |
|---|
| SMILES: | CC(OC1C(O)C(CO)OC(OP([O-])(=O)OP([O-])(=O)OCC2OC(C(O)C2O)N2C=CC(=O)NC2=O)C1NC(C)=O)C([O-])=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pyrimidine nucleotide sugars. These are pyrimidine nucleotides bound to a saccharide derivative through the terminal phosphate group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Pyrimidine nucleotides |
|---|
| Sub Class | Pyrimidine nucleotide sugars |
|---|
| Direct Parent | Pyrimidine nucleotide sugars |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyrimidine nucleotide sugar
- Pyrimidine ribonucleoside diphosphate
- Muramic_acid
- Pentose phosphate
- Pentose-5-phosphate
- N-acyl-alpha-hexosamine
- N-glycosyl compound
- Glycosyl compound
- Monosaccharide phosphate
- Organic pyrophosphate
- Pyrimidone
- Sugar acid
- Alkyl phosphate
- Phosphoric acid ester
- Oxane
- Hydropyrimidine
- Organic phosphoric acid derivative
- Pyrimidine
- Monosaccharide
- Heteroaromatic compound
- Acetamide
- Vinylogous amide
- Tetrahydrofuran
- Carboxamide group
- Lactam
- Urea
- Secondary carboxylic acid amide
- Secondary alcohol
- Carboxylic acid derivative
- Organoheterocyclic compound
- Azacycle
- Oxacycle
- Monocarboxylic acid or derivatives
- Ether
- Dialkyl ether
- Carboxylic acid
- Alcohol
- Carbonyl group
- Organonitrogen compound
- Organooxygen compound
- Primary alcohol
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Organic nitrogen compound
- Organic anion
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | -3 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | | Amino sugar and nucleotide sugar metabolism I | PW000886 | 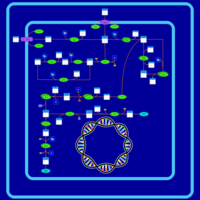   | | peptidoglycan biosynthesis I | PW000906 | 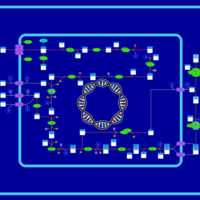 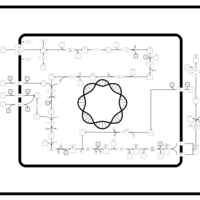 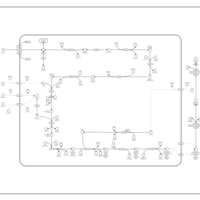 | | peptidoglycan biosynthesis I 2 | PW002062 | 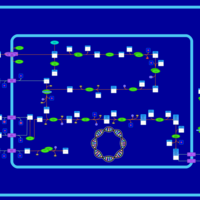 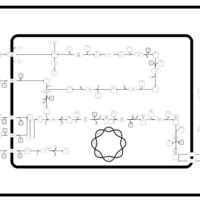 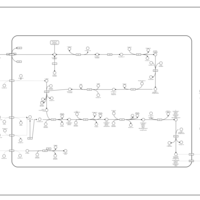 |
|
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | |
|---|
| References |
|---|
| References: | Not Available |
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | | Resource | Link |
|---|
| CHEBI ID | Not Available | | HMDB ID | Not Available | | Pubchem Compound ID | Not Available | | Kegg ID | Not Available | | ChemSpider ID | Not Available | | Wikipedia ID | Not Available | | BioCyc ID | Not Available |
|
|---|