Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
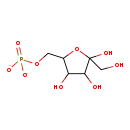
D-tagatofuranose 6-phosphate (M2MDB006322)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2015-09-08 17:50:04 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2016-09-13 16:35:44 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | D-tagatofuranose 6-phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | D-tagatofuranose 6-phosphate is an intermediate in galactitol degradation pathway in E.coli. It is a substrate for the enzyme 6-phosphofructokinase II which catalyzes the reaction D-tagatofuranose 6-phosphate + ATP -> D-tagatofuranose 1,6-bisphosphate + ADP + H+. It is also a product for enzyme galactitol-1-phosphate dehydrogenase which catalyzes reaction galactitol 1-phosphate + NAD+ -> D-tagatofuranose 6-phosphate + NADH + H+ (BioCyc compound: TAGATOSE-6-PHOSPHATE). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: Not Available Monoisotopic: Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | [3,4,5-trihydroxy-5-(hydroxymethyl)oxolan-2-yl]methyl phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | [3,4,5-trihydroxy-5-(hydroxymethyl)oxolan-2-yl]methyl phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as pentose phosphates. These are carbohydrate derivatives containing a pentose substituted by one or more phosphate groups. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic oxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Carbohydrates and carbohydrate conjugates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Pentose phosphates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic heteromonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | D-tagatofuranose 6-phosphate > β-D-glucose 6-phosphate β-D-glucose 6-phosphate + β-D-glucose 6-phosphate <> D-tagatofuranose 6-phosphate D-tagatofuranose 6-phosphate > beta-D-Glucose 6-phosphate Fructose 1,6-bisphosphate + Water + Fructose 1,6-bisphosphate > Phosphate + D-tagatofuranose 6-phosphate Galactose 1-phosphate + NAD + Galactose 1-phosphate > NADH + Hydrogen ion + D-tagatofuranose 6-phosphate D-tagatofuranose 6-phosphate + Adenosine triphosphate > Adenosine diphosphate + Hydrogen ion + D-tagatofuranose 1,6-bisphosphate + ADP α-D-mannose 6-phosphate <> D-tagatofuranose 6-phosphate D-tagatofuranose 6-phosphate > α-D-mannose 6-phosphate D-allulose 6-phosphate + D-Allulose-6-phosphate > D-tagatofuranose 6-phosphate Glucosamine 6-phosphate + Water > Ammonium + D-tagatofuranose 6-phosphate β-D-fructofuranose + Adenosine triphosphate + D-Fructose > Adenosine diphosphate + Hydrogen ion + D-tagatofuranose 6-phosphate + ADP D-Fructose + Adenosine triphosphate + D-Fructose > D-tagatofuranose 6-phosphate + Adenosine diphosphate + Hydrogen ion + ADP D-tagatofuranose 6-phosphate + L-Glutamine <> Glucosamine 6-phosphate + L-Glutamic acid + L-Glutamate β-D-fructofuranose + HPr - phosphorylated + D-Fructose > D-tagatofuranose 6-phosphate + HPr D-glucosamine 6-phosphate + Water > Ammonium + D-tagatofuranose 6-phosphate β-D-glucose 6-phosphate <> D-tagatofuranose 6-phosphate D-tagatofuranose 6-phosphate + L-Glutamine <> D-glucosamine 6-phosphate + L-Glutamate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in mannose-6-phosphate isomerase activity
- Specific function:
- Involved in the conversion of glucose to GDP-L-fucose, which can be converted to L-fucose, a capsular polysaccharide
- Gene Name:
- manA
- Uniprot ID:
- P00946
- Molecular weight:
- 42850
Reactions
| D-mannose 6-phosphate = D-fructose 6-phosphate. |
- General function:
- Involved in phosphotransferase activity, alcohol group as acceptor
- Specific function:
- ATP + D-fructose 6-phosphate = ADP + D- fructose 1,6-bisphosphate
- Gene Name:
- pfkB
- Uniprot ID:
- P06999
- Molecular weight:
- 32456
Reactions
| ATP + D-fructose 6-phosphate = ADP + D-fructose 1,6-bisphosphate. |
- General function:
- Involved in glucose-6-phosphate isomerase activity
- Specific function:
- D-glucose 6-phosphate = D-fructose 6- phosphate
- Gene Name:
- pgi
- Uniprot ID:
- P0A6T1
- Molecular weight:
- 61529
Reactions
| D-glucose 6-phosphate = D-fructose 6-phosphate. |
- General function:
- Involved in carbohydrate metabolic process
- Specific function:
- Catalyzes the reversible isomerization-deamination of glucosamine 6-phosphate (GlcN6P) to form fructose 6-phosphate (Fru6P) and ammonium ion
- Gene Name:
- nagB
- Uniprot ID:
- P0A759
- Molecular weight:
- 29774
Reactions
| D-glucosamine 6-phosphate + H(2)O = D-fructose 6-phosphate + NH(3). |
- General function:
- Involved in phosphoric ester hydrolase activity
- Specific function:
- D-fructose 1,6-bisphosphate + H(2)O = D- fructose 6-phosphate + phosphate
- Gene Name:
- fbp
- Uniprot ID:
- P0A993
- Molecular weight:
- 36834
Reactions
| D-fructose 1,6-bisphosphate + H(2)O = D-fructose 6-phosphate + phosphate. |
- General function:
- Involved in glycerol metabolic process
- Specific function:
- D-fructose 1,6-bisphosphate + H(2)O = D- fructose 6-phosphate + phosphate
- Gene Name:
- glpX
- Uniprot ID:
- P0A9C9
- Molecular weight:
- 35852
Reactions
| D-fructose 1,6-bisphosphate + H(2)O = D-fructose 6-phosphate + phosphate. |
- General function:
- Involved in zinc ion binding
- Specific function:
- Converts galactitol 1-phosphate to tagatose 6-phosphate
- Gene Name:
- gatD
- Uniprot ID:
- P0A9S3
- Molecular weight:
- 37390
Reactions
| Galactitol-1-phosphate + NAD(+) = L-tagatose 6-phosphate + NADH. |
- General function:
- Involved in metabolic process
- Specific function:
- Catalyzes the first step in hexosamine metabolism, converting fructose-6P into glucosamine-6P using glutamine as a nitrogen source
- Gene Name:
- glmS
- Uniprot ID:
- P17169
- Molecular weight:
- 66894
Reactions
| L-glutamine + D-fructose 6-phosphate = L-glutamate + D-glucosamine 6-phosphate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the dephosphorylation of the artificial chromogenic substrate p-nitrophenyl phosphate (pNPP) and of the natural substrates pyridoxalphosphate and erythrose 4-phosphate
- Gene Name:
- ybhA
- Uniprot ID:
- P21829
- Molecular weight:
- 30201
- General function:
- Transcription
- Specific function:
- Catalyzes the phosphorylation of fructose to fructose-6- P. Has also low level glucokinase activity in vitro. Is not able to phosphorylate D-ribose, D-mannitol, D-sorbitol, inositol, and L-threonine
- Gene Name:
- mak
- Uniprot ID:
- P23917
- Molecular weight:
- 32500
Reactions
| ATP + D-fructose = ADP + D-fructose 6-phosphate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Specific function unknown
- Gene Name:
- alsE
- Uniprot ID:
- P32719
- Molecular weight:
- 26109
- General function:
- Involved in phosphoenolpyruvate-dependent sugar phosphotransferase system
- Specific function:
- The phosphoenolpyruvate-dependent sugar phosphotransferase system (sugar PTS), a major carbohydrate active -transport system, catalyzes the phosphorylation of incoming sugar substrates concomitantly with their translocation across the cell membrane. This system is involved in mannose transport
- Gene Name:
- manX
- Uniprot ID:
- P69797
- Molecular weight:
- 35047
Reactions
| Protein EIIA N(pi)-phospho-L-histidine + protein EIIB = protein EIIA + protein EIIB N(pi)-phospho-L-histidine/cysteine. |
| Protein EIIB N(pi)-phospho-L-histidine/cysteine + sugar = protein EIIB + sugar phosphate. |
- General function:
- Involved in phosphoenolpyruvate-dependent sugar phosphotransferase system
- Specific function:
- The phosphoenolpyruvate-dependent sugar phosphotransferase system (PTS), a major carbohydrate active -transport system, catalyzes the phosphorylation of incoming sugar substrates concomitant with their translocation across the cell membrane. This system is involved in mannose transport
- Gene Name:
- manY
- Uniprot ID:
- P69801
- Molecular weight:
- 27636
- General function:
- Involved in phosphoenolpyruvate-dependent sugar phosphotransferase system
- Specific function:
- The phosphoenolpyruvate-dependent sugar phosphotransferase system (PTS), a major carbohydrate active -transport system, catalyzes the phosphorylation of incoming sugar substrates concomitant with their translocation across the cell membrane. This system is involved in mannose transport
- Gene Name:
- manZ
- Uniprot ID:
- P69805
- Molecular weight:
- 31303
- General function:
- Involved in glycerol metabolic process
- Specific function:
- Specific function unknown
- Gene Name:
- yggF
- Uniprot ID:
- P21437
- Molecular weight:
- 34323
Reactions
| D-fructose 1,6-bisphosphate + H(2)O = D-fructose 6-phosphate + phosphate. |
Transporters
- General function:
- Involved in phosphoenolpyruvate-dependent sugar phosphotransferase system
- Specific function:
- The phosphoenolpyruvate-dependent sugar phosphotransferase system (PTS), a major carbohydrate active -transport system, catalyzes the phosphorylation of incoming sugar substrates concomitant with their translocation across the cell membrane. This system is involved in mannose transport
- Gene Name:
- manY
- Uniprot ID:
- P69801
- Molecular weight:
- 27636
- General function:
- Involved in phosphoenolpyruvate-dependent sugar phosphotransferase system
- Specific function:
- The phosphoenolpyruvate-dependent sugar phosphotransferase system (PTS), a major carbohydrate active -transport system, catalyzes the phosphorylation of incoming sugar substrates concomitant with their translocation across the cell membrane. This system is involved in mannose transport
- Gene Name:
- manZ
- Uniprot ID:
- P69805
- Molecular weight:
- 31303