| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2015-06-04 15:16:17 -0600 |
|---|
| Update Date | 2015-07-23 12:07:06 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | L-Glutamyl-tRNA(Glu) |
|---|
| Description | |
|---|
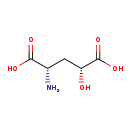
| Structure | |
|---|
| Synonyms: | - (2S,4R)-2-amino-4-Hydroxypentanedioate
- (2S,4R)-2-amino-4-Hydroxypentanedioic acid
- L-erythro-4-Hydroxyglutamate
- L-erythro-4-Hydroxyglutamic acid
|
|---|
| Chemical Formula: | C5H9NO5 |
|---|
| Weight: | Average: 163.1287
Monoisotopic: 163.048072403 |
|---|
| InChI Key: | HBDWQSHEVMSFGY-STHAYSLISA-N |
|---|
| InChI: | InChI=1S/C5H9NO5/c6-2(4(8)9)1-3(7)5(10)11/h2-3,7H,1,6H2,(H,8,9)(H,10,11)/t2-,3+/m0/s1 |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | (2S,4R)-2-amino-4-hydroxypentanedioic acid |
|---|
| Traditional IUPAC Name: | (2S,4R)-2-amino-4-hydroxypentanedioic acid |
|---|
| SMILES: | N[C@@H](C[C@@H](O)C(O)=O)C(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glutamic acid and derivatives. Glutamic acid and derivatives are compounds containing glutamic acid or a derivative thereof resulting from reaction of glutamic acid at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Glutamic acid and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glutamic acid or derivatives
- Alpha-amino acid
- L-alpha-amino acid
- Amino fatty acid
- Short-chain hydroxy acid
- Hydroxy fatty acid
- Alpha-hydroxy acid
- Fatty acyl
- Fatty acid
- Dicarboxylic acid or derivatives
- Hydroxy acid
- 1,3-aminoalcohol
- Amino acid
- Secondary alcohol
- Carboxylic acid
- Organic nitrogen compound
- Primary aliphatic amine
- Organonitrogen compound
- Organooxygen compound
- Primary amine
- Hydrocarbon derivative
- Carbonyl group
- Organic oxide
- Amine
- Organopnictogen compound
- Organic oxygen compound
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | -1 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00dl-9300000000-192385d58efcfe6d927a | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0fr2-1900000000-1952d82430e8d4d017ec | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0g4i-8900000000-9b552b5028c7cac0636c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-006x-9200000000-69e6a42e7b8c893be2b0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-3900000000-a1ac74b59a8487175409 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00di-9600000000-b341efd7d7ac7e6a7652 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00di-9100000000-a0987d89d6b50b5ee46a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0gi0-3900000000-72a434ff0c346ae54bf5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0fk9-9600000000-16a8930ac7a8ed9a8656 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00di-9100000000-4825e58f647c6709f63e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0900000000-f33be61e6a7e963f5725 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0079-9200000000-d4c3022fe2d2d99ba90b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-ce7c939afbade242bd22 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | Not Available |
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|