| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-11-06 04:55:28 -0700 |
|---|
| Update Date | 2015-06-03 17:27:29 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
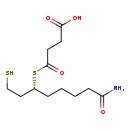
| Name: | (S)-Succinyldihydrolipoamide |
|---|
| Description | (S)-Succinyldihydrolipoamide is a metabolite (a product as well as a substrate) in glutamate degradation. It is also invovled in citrate cycle and is a reactant for oxoglutarate dehydrogenase (EC 1.2.4.2) and dihydrolipoyllysine-residue succinyltransferase (EC 2.3.1.61). |
|---|
| Structure | |
|---|
| Synonyms: | - 4-[(3R)-8-amino-8-oxo-1-Sulfanyloctan-3-yl]sulfanyl-4-oxobutanoate
- 4-[(3R)-8-amino-8-oxo-1-sulfanyloctan-3-yl]sulfanyl-4-oxobutanoic acid
- 4-[(3R)-8-amino-8-oxo-1-Sulphanyloctan-3-yl]sulphanyl-4-oxobutanoate
- 4-[(3R)-8-amino-8-oxo-1-Sulphanyloctan-3-yl]sulphanyl-4-oxobutanoic acid
- S-succinyl-dihydrolipoamide
- S-Succinyldihydrolipoamide
|
|---|
| Chemical Formula: | C12H21NO4S2 |
|---|
| Weight: | Average: 307.429
Monoisotopic: 307.091199545 |
|---|
| InChI Key: | RJCJWONCSKSHES-SECBINFHSA-N |
|---|
| InChI: | InChI=1S/C12H21NO4S2/c13-10(14)4-2-1-3-9(7-8-18)19-12(17)6-5-11(15)16/h9,18H,1-8H2,(H2,13,14)(H,15,16)/t9-/m1/s1 |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | 4-{[(3R)-7-carbamoyl-1-sulfanylheptan-3-yl]sulfanyl}-4-oxobutanoic acid |
|---|
| Traditional IUPAC Name: | 4-{[(3R)-7-carbamoyl-1-sulfanylheptan-3-yl]sulfanyl}-4-oxobutanoic acid |
|---|
| SMILES: | NC(=O)CCCC[C@H](CCS)SC(=O)CCC(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as thia fatty acids. These are fatty acid derivatives obtained by insertion of a sulfur atom at specific positions in the chain. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acids and conjugates |
|---|
| Direct Parent | Thia fatty acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Thia fatty acid
- Fatty acyl thioester
- Fatty amide
- Carboxamide group
- Primary carboxylic acid amide
- Thiocarboxylic acid ester
- Carbothioic s-ester
- Sulfenyl compound
- Thiocarboxylic acid or derivatives
- Alkylthiol
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Carbonyl group
- Organic oxygen compound
- Organosulfur compound
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Organopnictogen compound
- Hydrocarbon derivative
- Organic oxide
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | -1 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | | glycolysis and pyruvate dehydrogenase | PW000785 | 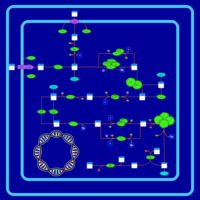   |
|
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0ukc-9880000000-d66345c90ce5eb67fa60 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00dl-9533000000-a1ebe0f3e16866573001 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0596-0190000000-b49046dab0850898c889 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-006y-2190000000-474bcff100b398100e49 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0nt9-4910000000-247a7886ecdb5a884d7c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0596-0190000000-b49046dab0850898c889 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-006y-2190000000-474bcff100b398100e49 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0nt9-4910000000-247a7886ecdb5a884d7c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-5294000000-672a48ef8e666cb0ee5f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4j-9360000000-f9e4f05adcb51d48447c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-000x-9100000000-d7f410ba584f3aad89cc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-1229000000-d6ef878a291e343f754a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-7970000000-eff246297f1390d8e037 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-000x-9500000000-66fb2d1c65f8ee705da4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-0369000000-035798c3620974bb7aab | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03dl-1910000000-e3af53d75b9037036cfe | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udr-5900000000-f70f4c6e3a3b250e485f | View in MoNA |
|---|
|
|---|
| References |
|---|
| References: | Not Available |
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|