| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-11-06 04:55:27 -0700 |
|---|
| Update Date | 2015-06-03 17:27:28 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
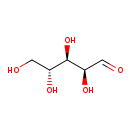
| Name: | D-Arabinose |
|---|
| Description | Arabinose is an aldopentose – a monosaccharide containing five carbon atoms, and including an aldehyde (CHO) functional group. (Wikipedia) D-Arabinose is degraded by Escherichia coli B via some of the L-fucose pathway enzymes and a D-ribulokinase which is distinct from the L-fuculokinase of the L-fucose pathway. (PMID 3056899) |
|---|
| Structure | |
|---|
| Synonyms: | - aldehydo-D-arabinose
- Arabinose
|
|---|
| Chemical Formula: | C5H10O5 |
|---|
| Weight: | Average: 150.1299
Monoisotopic: 150.05282343 |
|---|
| InChI Key: | PYMYPHUHKUWMLA-WDCZJNDASA-N |
|---|
| InChI: | InChI=1S/C5H10O5/c6-1-3(8)5(10)4(9)2-7/h1,3-5,7-10H,2H2/t3-,4-,5+/m1/s1 |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | (2S,3R,4R)-2,3,4,5-tetrahydroxypentanal |
|---|
| Traditional IUPAC Name: | arabinose |
|---|
| SMILES: | OC[C@@H](O)[C@@H](O)[C@H](O)C=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pentoses. These are monosaccharides in which the carbohydrate moiety contains five carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Pentoses |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pentose monosaccharide
- Beta-hydroxy aldehyde
- Alpha-hydroxyaldehyde
- Secondary alcohol
- Polyol
- Organic oxide
- Hydrocarbon derivative
- Short-chain aldehyde
- Primary alcohol
- Carbonyl group
- Aldehyde
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | 0 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-052f-9100000000-3d844ea1b588172feaf0 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-05i0-6249400000-6324b88618b9174f1ee5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , negative | splash10-08mi-9100000000-fdc6cb3672d0fd9782e4 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF 10V, negative | splash10-0a4i-9000000000-8b4cbdfdbd1d64750afc | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - QqQ 1V, negative | splash10-0002-1900000000-a6fad2b086e4d72e6455 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - QqQ 4V, negative | splash10-000j-9500000000-7f3c671f5a2325c13521 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - QqQ 8V, negative | splash10-052r-9000000000-4607894e05698b4efd4b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - QqQ 10V, negative | splash10-0a4r-9000000000-7713ba3bfd41873ed05b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - QqQ 14V, negative | splash10-0a4i-9000000000-19cc19a6070f8904f223 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - QqQ 17V, negative | splash10-0a4i-9000000000-cbfe90fac8fd1f09d178 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - QqQ 20V, negative | splash10-0a4i-9000000000-194cea61e7f4bb116108 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 10V, negative | splash10-001i-2900000000-0431fbb715edbb5c2854 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 0V, negative | splash10-001i-0900000000-9b7382416823533b7db1 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 0V, negative | splash10-001i-0900000000-b354fa776d3c927b52a8 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 0V, negative | splash10-001i-1900000000-4f54308f655454dcf238 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 1V, negative | splash10-001i-2900000000-ffb4e5e554a72f34fd7b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 1V, negative | splash10-001i-4900000000-df24839373685cc97976 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 2V, negative | splash10-001r-7900000000-08514302c6b21ad9497c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 2V, negative | splash10-008i-9700000000-9b73d2467b158cb7fe64 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 3V, negative | splash10-00dr-9300000000-80b57725a8299b2e9afa | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 4V, negative | splash10-00dr-9100000000-f5eb0b039697ddead9f8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-2900000000-f8b9c6d77ac7485b2a13 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-9300000000-c381cba09705c9d22367 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-08fv-9000000000-157eda9875c5cd43b332 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000j-9400000000-8bb17d4c0bc5362b2610 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-06ri-9200000000-f20b8cc45d0196859991 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9000000000-83841c4e2cbd5aa6df09 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | Not Available |
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|