| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-10-10 12:22:30 -0600 |
|---|
| Update Date | 2015-09-16 14:50:11 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | N-Acetyl-alpha-neuraminate |
|---|
| Description | N-Acetyl-alpha-neuraminate is a sialic acid. Sialic acids are an important family of related 9-carbon sugars acids, present on the surface of many different cells and functioning in a wide range of different biological processes. They can be utilized by pathogens to evade the host immune response. N-acetylneuraminic acid is the most common sialic acid. A number of bacteria that can colonize humans (such as E. coli) make use of N-acetylneuraminic acid as a nutrient source. Production of the enzymes involved in N-acetylneuraminate degradation is induced by growth on sialic acid as the sole source of carbon, which is due to transcriptional regulation by the repressor NanR.
|
|---|
| Structure | |
|---|
| Synonyms: | - (2R,4S,5R,6R)-5-acetamido-2,4-Dihydroxy-6-(1R,2R)-1,2,3-trihydroxypropyloxane-2-carboxylate
- (2R,4S,5R,6R)-5-acetamido-2,4-dihydroxy-6-(1R,2R)-1,2,3-trihydroxypropyloxane-2-carboxylic acid
- 5-(acetylamino)-3,5-dideoxy-D-glycero-a-D-galacto-2-Nonulopyranosonate
- 5-(acetylamino)-3,5-dideoxy-D-glycero-a-D-galacto-2-Nonulopyranosonic acid
- 5-(acetylamino)-3,5-Dideoxy-delta-glycero-a-delta-galacto-2-nonulopyranosonate
- 5-(acetylamino)-3,5-Dideoxy-delta-glycero-a-delta-galacto-2-nonulopyranosonic acid
- 5-(acetylamino)-3,5-dideoxy-delta-glycero-alpha-delta-galacto-2-Nonulopyranosonate
- 5-(acetylamino)-3,5-dideoxy-delta-glycero-alpha-delta-galacto-2-Nonulopyranosonic acid
- 5-(acetylamino)-3,5-Dideoxy-δ-glycero-a-δ-galacto-2-nonulopyranosonate
- 5-(acetylamino)-3,5-Dideoxy-δ-glycero-a-δ-galacto-2-nonulopyranosonic acid
- 5-(acetylamino)-3,5-Dideoxy-δ-glycero-α-δ-galacto-2-nonulopyranosonate
- 5-(acetylamino)-3,5-Dideoxy-δ-glycero-α-δ-galacto-2-nonulopyranosonic acid
- 5-acetamido-3,5-dideoxy-a-D-glycero-D-galacto-Nonulopyranosonate
- 5-acetamido-3,5-dideoxy-a-D-glycero-D-galacto-Nonulopyranosonic acid
- 5-acetamido-3,5-Dideoxy-a-delta-glycero-delta-galacto-nonulopyranosonate
- 5-acetamido-3,5-Dideoxy-a-delta-glycero-delta-galacto-nonulopyranosonic acid
- 5-acetamido-3,5-Dideoxy-a-δ-glycero-δ-galacto-nonulopyranosonate
- 5-acetamido-3,5-Dideoxy-a-δ-glycero-δ-galacto-nonulopyranosonic acid
- 5-acetamido-3,5-dideoxy-alpha-delta-glycero-delta-galacto-Nonulopyranosonate
- 5-acetamido-3,5-dideoxy-alpha-delta-glycero-delta-galacto-Nonulopyranosonic acid
- 5-acetamido-3,5-Dideoxy-α-δ-glycero-δ-galacto-nonulopyranosonate
- 5-acetamido-3,5-Dideoxy-α-δ-glycero-δ-galacto-nonulopyranosonic acid
- 8)-a-NeuNAC-(2->n
- 8)-alpha-NeuNAC-(2->n
- 8)-α-NeuNAC-(2->n
- a-Neu5ac
- a-Neu5ac-(2->8)N
- Alpha-Neu5Ac
- Alpha-Neu5Ac-(2->8)n
- N-Acetyl-a-D-neuraminate
- N-Acetyl-a-D-neuraminic acid
- N-Acetyl-a-delta-neuraminate
- N-Acetyl-a-delta-neuraminic acid
- N-Acetyl-a-neuraminate
- N-Acetyl-a-neuraminic acid
- N-Acetyl-a-δ-neuraminate
- N-Acetyl-a-δ-neuraminic acid
- N-Acetyl-alpha-delta-neuraminate
- N-Acetyl-alpha-delta-neuraminic acid
- N-Acetyl-alpha-neuraminic acid
- N-Acetyl-α-neuraminate
- N-Acetyl-α-neuraminic acid
- N-Acetyl-α-δ-neuraminate
- N-Acetyl-α-δ-neuraminic acid
- N-Acetylated a-(2->8)-linked homosialopolysaccharide
- N-acetylated alpha-(2->8)-linked homosialopolysaccharide
- N-Acetylated α-(2->8)-linked homosialopolysaccharide
- Poly-a-(2->8)-neu5ac
- Poly-a-(2->8)-neunac
- Poly-alpha-(2->8)-Neu5Ac
- Poly-alpha-(2->8)-NeuNAc
- Poly-α-(2->8)-neu5ac
- Poly-α-(2->8)-neunac
- Polyalpha-Neu5Ac-(2->8)
- α-Neu5ac
- α-Neu5ac-(2->8)N
|
|---|
| Chemical Formula: | C11H19NO9 |
|---|
| Weight: | Average: 309.2699
Monoisotopic: 309.105981211 |
|---|
| InChI Key: | SQVRNKJHWKZAKO-YRMXFSIDSA-N |
|---|
| InChI: | InChI=1S/C11H19NO9/c1-4(14)12-7-5(15)2-11(20,10(18)19)21-9(7)8(17)6(16)3-13/h5-9,13,15-17,20H,2-3H2,1H3,(H,12,14)(H,18,19)/t5-,6+,7+,8+,9+,11+/m0/s1 |
|---|
| CAS number: | 21646-00-4 |
|---|
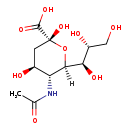
| IUPAC Name: | (2R,4S,5R,6R)-5-acetamido-2,4-dihydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]oxane-2-carboxylic acid |
|---|
| Traditional IUPAC Name: | N-acetylneuraminic acid |
|---|
| SMILES: | [H][C@]1(O[C@](O)(C[C@H](O)[C@H]1NC(C)=O)C(O)=O)[C@H](O)[C@H](O)CO |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as n-acylneuraminic acids. These are neuraminic acids carrying an N-acyl substituent. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | N-acylneuraminic acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - N-acylneuraminic acid
- Neuraminic acid
- C-glucuronide
- C-glycosyl compound
- Glycosyl compound
- Alpha-hydroxy acid
- Pyran
- Hydroxy acid
- Oxane
- Acetamide
- Carboxamide group
- Hemiacetal
- Secondary carboxylic acid amide
- Secondary alcohol
- Carboxylic acid derivative
- Carboxylic acid
- Oxacycle
- Organoheterocyclic compound
- Monocarboxylic acid or derivatives
- Polyol
- Alcohol
- Hydrocarbon derivative
- Organic nitrogen compound
- Organopnictogen compound
- Primary alcohol
- Organic oxide
- Carbonyl group
- Organonitrogen compound
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -1 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-03dl-9370000000-a2a8e1f2580c7bb97655 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (5 TMS) - 70eV, Positive | splash10-0pb9-2390148000-6c04c73ad5de1e54ba0e | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_5_15) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("N-Acetyl-a-neuraminic acid,5TBDMS,#15" TMS) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01ox-1092000000-ca2f76604846382a3032 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03kc-6190000000-7e304e9fa3125067d352 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-08fu-9240000000-65d03a6c6f60a71416c6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-074j-4961000000-bc80694ae9687db9a365 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-9210000000-c1d9f41a64bc464147dd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9300000000-a1d857dc5ad7118647b3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03kc-0095000000-8f7746cc3f652b12033f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-02tc-1392000000-3e251c9d2583e71f866a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03di-7910000000-ba41b78e53cb934a9b19 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-3039000000-4348eff0515b0da3bc08 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a59-6290000000-5ac6f13cca0827ab0dd0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4u-9100000000-ac22477eeca403481eff | View in MoNA |
|---|
|
|---|
| References |
|---|
| References: | - Thompson JA, Miles BS, Fennessey PV: Urinary organic acids quantitated by age groups in a healthy pediatric population. Clin Chem. 1977 Sep;23(9):1734-8. Pubmed: 890917
|
|---|
| Synthesis Reference: | Baumberger, Franz; Vasella, Andrea. , Synthesis of N-acetylneuraminic acid and N-acetyl-4-epineuraminic acid. Helvetica Chimica Acta (1986), 69(5), 1205-15. |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|