| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-10-10 12:15:07 -0600 |
|---|
| Update Date | 2015-06-03 17:25:52 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | L-Rhamnose |
|---|
| Description | Rhamnose (Rham) is a naturally occurring deoxy sugar. It can be classified as either a methyl-pentose or a 6-deoxy-hexose. Rhamnose occurs in nature in its L-form as L-rhamnose (6-deoxy-L-mannose). This is unusual, since most of the naturally occurring sugars are in D-form. Rhamnose is commonly bound to other sugars in nature. It is a common glycone component of glycosides from many plants. Rhamnose is also a component of the outer cell membrane of certain bacteria. L-rhamnose is metabolized to L-Lactaldehyde, which is a branching point in the metabolic pathway of L-fucose and L-rhamnose utilization. It exists in two anomeric forms, alpha-L-rhamnose and beta-L-rhamnose. |
|---|
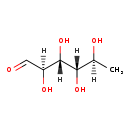
| Structure | |
|---|
| Synonyms: | - 6-Deoxy-L(+)-mannose
- 6-Deoxy-L-Mannopyranose
- 6-Deoxy-L-mannose
- 6-deoxy-Mannose
- 6-Deoxyhexopyranose
- 6-deoxyhexose
- 6-Deoxymannose
- Isodulcit
- Isodulcitol
- L(+)-Rhamnose
- L(+)-Rhamnose monohydrate
- L(+)-Rhamnose monohydric acid
- L-(+)-Rhamnose
- L-(+)-Rhamnose 1-hydrate
- L-(+)-Rhamnose 1-hydric acid
- L-(+)-Rhamnose Hydrate = 6-Deoxy-L-mannose Monohydrate
- L-(+)-Rhamnose hydric acid = 6-deoxy-L-mannose monohydric acid
- L-(+)-Rhamnose monohydrate
- L-(+)-Rhamnose monohydric acid
- L-Mannomethylose
- L-Rha
- L-Rhamnopyranose
- L-rhamnose for biochemistry
- L-Rhamnose Monohydrate
- L-Rhamnose monohydric acid
- Locaose
- Mono(6-deoxy-b-L-mannopyranosyl) ester
- Mono(6-deoxy-beta-L-mannopyranosyl) ester
- mono(6-Deoxy-β-L-mannopyranosyl) ester
- P-(6-deoxy-b-L-mannopyranosyl) ester
- P-(6-deoxy-beta-L-mannopyranosyl) ester
- P-(6-Deoxy-β-L-mannopyranosyl) ester
- Rhamnopyranose
- Rhamnose
- Rhamnosemonohydrate
- Rhamnosemonohydric acid
- UDP-rhamnose
- Uridine 5-pyrophosphate
- Uridine 5-pyrophosphoric acid
- Uridine diphosphate rhamnose
- Uridine diphosphoric acid rhamnose
|
|---|
| Chemical Formula: | C6H12O5 |
|---|
| Weight: | Average: 164.1565
Monoisotopic: 164.068473494 |
|---|
| InChI Key: | PNNNRSAQSRJVSB-BXKVDMCESA-N |
|---|
| InChI: | InChI=1S/C6H12O5/c1-3(8)5(10)6(11)4(9)2-7/h2-6,8-11H,1H3/t3-,4-,5-,6-/m0/s1 |
|---|
| CAS number: | 3615-41-6 |
|---|
| IUPAC Name: | (2R,3R,4S,5S)-2,3,4,5-tetrahydroxyhexanal |
|---|
| Traditional IUPAC Name: | 6-deoxy-L-mannose |
|---|
| SMILES: | [H][C@@](C)(O)[C@]([H])(O)[C@@]([H])(O)[C@@]([H])(O)C=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as hexoses. These are monosaccharides in which the sugar unit is a is a six-carbon containing moeity. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Hexoses |
|---|
| Alternative Parents | |
|---|
| Substituents | - Hexose monosaccharide
- Medium-chain aldehyde
- Beta-hydroxy aldehyde
- Alpha-hydroxyaldehyde
- Secondary alcohol
- Polyol
- Organic oxide
- Hydrocarbon derivative
- Carbonyl group
- Aldehyde
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | 0 |
|---|
| Melting point: | 122 C |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | - Fructose and mannose metabolism ec00051
- Microbial metabolism in diverse environments ec01120
|
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (1 MEOX; 4 TMS) | splash10-014i-0920000000-3f32654cd6b207d5192c | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (1 MEOX; 4 TMS) | splash10-014i-0910000000-3efed9ba21b4acc1a0a7 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-014i-0920000000-15cadd38d19e98599380 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-014i-0920000000-845777ce5ccb02564dd4 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-014i-0910000000-a3998948f1f154365fea | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-014i-0900000000-09ff33efc7ead61776ac | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-000i-0900000000-c17a9df600308d9b31be | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF 10V, negative | splash10-0a4i-9300000000-aba9c0e75315919d15ca | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - QqQ 2V, negative | splash10-03di-0900000000-5bc4a1e81d1743e4d888 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - QqQ 6V, negative | splash10-114r-4900000000-cce3aaf678cc70d6861a | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - QqQ 8V, negative | splash10-0pb9-9700000000-4f6230f5365dba6dd52f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - QqQ 12V, negative | splash10-0a4i-9100000000-9477982822aed3518682 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - QqQ 16V, negative | splash10-0a4i-9000000000-594366ab16c0cac99671 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - QqQ 20V, negative | splash10-0a4i-9000000000-f0b38b5601df3611821c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 11V, negative | splash10-0udi-3900000000-a5276b0d1c6f7703894f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 11V, negative | splash10-004i-2900000000-2a20cb5f730a3fd74448 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 11V, negative | splash10-0089-9000000000-643fd0369e6935a5767c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 11V, negative | splash10-0532-9400000000-2e4332a61dcaf8add5aa | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014j-3900000000-ff3539fb021325c23c4a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a6r-9200000000-b270b5d83d44008211ba | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9000000000-f516852459b277fdb157 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0ly9-9600000000-efc90d58c6475abfcecd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0abi-9100000000-3b89500afd7b488df6fa | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9000000000-7b10fe17f3ce9401fd3f | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | Not Available |
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|