| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-10-10 12:13:45 -0600 |
|---|
| Update Date | 2015-06-03 17:25:48 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
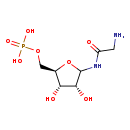
| Name: | 5'-Phospho-ribosylglycinamide |
|---|
| Description | 5-Phospho-ribosylglycinamide is an intermediate in puriine biosynthesis. It is a substrate for the enzyme Phosphoribosylglycinamide synthetase (EC:6.3.4.13) (GARS) which catalyses the second step in the de novo biosynthesis of purine. The reaction catalysed by phosphoribosylglycinamide synthetase is the ATP-dependent addition of 5-phosphoribosylamine to glycine to form 5'-phosphoribosylglycinamide. |
|---|
| Structure | |
|---|
| Synonyms: | - (2R,3S,4R)-5-(2-aminoacetyl)amino-3,4-dihydroxyoxolan-2-ylmethyl dihydrogen phosphate
- (2R,3S,4R)-5-(2-Aminoacetyl)amino-3,4-dihydroxyoxolan-2-ylmethyl dihydrogen phosphoric acid
- 5'-Phosphoribosylglycinamide
- 5'-Phosphoribosylglycineamide
- GAR
- Glycinamide ribonucleotide
- Glycineamide ribonucleotide
- Glycineamideribotide
- N-glycyl-5-O-phosphono-D-ribofuranosylamine
- N1-(5-Phospho-D-ribosyl)glycinamide
|
|---|
| Chemical Formula: | C7H15N2O8P |
|---|
| Weight: | Average: 286.1764
Monoisotopic: 286.056601978 |
|---|
| InChI Key: | OBQMLSFOUZUIOB-HJZCUYRDSA-N |
|---|
| InChI: | InChI=1S/C7H15N2O8P/c8-1-4(10)9-7-6(12)5(11)3(17-7)2-16-18(13,14)15/h3,5-7,11-12H,1-2,8H2,(H,9,10)(H2,13,14,15)/t3-,5-,6-,7?/m1/s1 |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | {[(2R,3S,4R)-5-(2-aminoacetamido)-3,4-dihydroxyoxolan-2-yl]methoxy}phosphonic acid |
|---|
| Traditional IUPAC Name: | 5'-phosphoribosylglycinamide |
|---|
| SMILES: | NCC(=O)NC1O[C@H](COP(O)(O)=O)[C@@H](O)[C@H]1O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glycinamide ribonucleotides. Glycinamide ribonucleotides are compounds in which the amide N atom of glycineamide is linked to the C-1 of a ribosyl (or deoxyribosyl) moiety. Nucleotides have a phosphate group linked to the C5 carbon of the ribose (or deoxyribose) moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Glycinamide ribonucleotides |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Glycinamide ribonucleotides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glycinamide-ribonucleotide
- Pentose phosphate
- Pentose-5-phosphate
- Alpha-amino acid amide
- Alpha-amino acid or derivatives
- Monosaccharide phosphate
- Pentose monosaccharide
- Monoalkyl phosphate
- Monosaccharide
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Alkyl phosphate
- Tetrahydrofuran
- Secondary carboxylic acid amide
- Secondary alcohol
- Amino acid or derivatives
- Carboxamide group
- 1,2-diol
- Organoheterocyclic compound
- Oxacycle
- Carboxylic acid derivative
- Organic nitrogen compound
- Organic oxide
- Primary aliphatic amine
- Organopnictogen compound
- Alcohol
- Organonitrogen compound
- Carbonyl group
- Organooxygen compound
- Amine
- Organic oxygen compound
- Hydrocarbon derivative
- Primary amine
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | -1 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | |
|---|
| References |
|---|
| References: | Not Available |
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | | Resource | Link |
|---|
| CHEBI ID | 18349 | | HMDB ID | Not Available | | Pubchem Compound ID | 5459918 | | Kegg ID | Not Available | | ChemSpider ID | Not Available | | Wikipedia ID | Not Available | | BioCyc ID | Not Available |
|
|---|