| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-08-09 09:25:24 -0600 |
|---|
| Update Date | 2015-06-03 17:21:45 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | L-Histidinal |
|---|
| Description | L-histidinal is a member of the chemical class known as Imidazoles. These are compounds containing an imidazole ring, which is an aromatic five-member ring with two nitrogen atoms at positions 1 and 3, and three carbon atoms. L-histidinal is invovled in Biosynthesis of secondary metabolites, and Histidine metabolism. (KEGG) |
|---|
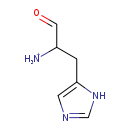
| Structure | |
|---|
| Synonyms: | |
|---|
| Chemical Formula: | C6H10N3O |
|---|
| Weight: | Average: 140.1631
Monoisotopic: 140.082386957 |
|---|
| InChI Key: | VYOIELONWKIZJS-UHFFFAOYSA-O |
|---|
| InChI: | InChI=1S/C6H9N3O/c7-5(3-10)1-6-2-8-4-9-6/h2-5H,1,7H2,(H,8,9)/p+1 |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | 1-(1H-imidazol-5-yl)-3-oxopropan-2-aminium |
|---|
| Traditional IUPAC Name: | 1-(3H-imidazol-4-yl)-3-oxopropan-2-aminium |
|---|
| SMILES: | [NH3+]C(CC1=CN=CN1)C=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as aralkylamines. These are alkylamines in which the alkyl group is substituted at one carbon atom by an aromatic hydrocarbyl group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic nitrogen compounds |
|---|
| Class | Organonitrogen compounds |
|---|
| Sub Class | Amines |
|---|
| Direct Parent | Aralkylamines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Aralkylamine
- Heteroaromatic compound
- Imidazole
- Azole
- Azacycle
- Organoheterocyclic compound
- Organic oxygen compound
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Primary amine
- Organooxygen compound
- Primary aliphatic amine
- Carbonyl group
- Aldehyde
- Organic cation
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | 1 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | |
|---|
| References |
|---|
| References: | - Winder, C. L., Dunn, W. B., Schuler, S., Broadhurst, D., Jarvis, R., Stephens, G. M., Goodacre, R. (2008). "Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites." Anal Chem 80:2939-2948. Pubmed: 18331064
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | | Resource | Link |
|---|
| CHEBI ID | Not Available | | HMDB ID | Not Available | | Pubchem Compound ID | 25244065 | | Kegg ID | C01929 | | ChemSpider ID | 3824841 | | Wikipedia ID | Not Available | | BioCyc ID | Not Available |
|
|---|