| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-08-09 09:16:19 -0600 |
|---|
| Update Date | 2015-06-03 17:21:33 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | Pyrroloquinoline quinone |
|---|
| Description | Enzymes containing PQQ are called quinoproteins. PQQ and quinoproteins play a role in the redox metabolism and structural integrity of cells and tissues [PMID:2558842]. It was reported that aminoadipate semialdehyde dehydrogenase (AASDH) might also use PQQ as a cofactor, suggesting a possibility that PQQ is a vitamin in mammals. [PMID:12712191] |
|---|
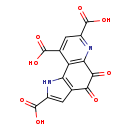
| Structure | |
|---|
| Synonyms: | - 2,7,9-Tricarboxy-1H-pyrrolo[2,3-f]quinoline-4,5-dione
- Methoxatin
- PQQ
- Pyrrolo-quinoline quinone
- Pyrroloquinoline-quinone
|
|---|
| Chemical Formula: | C14H6N2O8 |
|---|
| Weight: | Average: 330.206
Monoisotopic: 330.012415178 |
|---|
| InChI Key: | MMXZSJMASHPLLR-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/C14H6N2O8/c17-10-4-2-6(14(23)24)15-8(4)7-3(12(19)20)1-5(13(21)22)16-9(7)11(10)18/h1-2,15H,(H,19,20)(H,21,22)(H,23,24) |
|---|
| CAS number: | 72909-34-3 |
|---|
| IUPAC Name: | 4,5-dioxo-1H,4H,5H-pyrrolo[2,3-f]quinoline-2,7,9-tricarboxylic acid |
|---|
| Traditional IUPAC Name: | pyrroloquinoline quinone |
|---|
| SMILES: | OC(=O)C1=CC2=C(N1)C1=C(N=C(C=C1C(O)=O)C(O)=O)C(=O)C2=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pyrroloquinoline quinones. Pyrroloquinoline quinones are compounds with a structure based on the 2,7,-tricarboxy-1H-pyrrolo[2,3-f ]quinoline-4,5-dione. Pyrroloquinoline Quinones usually bear a carboxylic acid group at the C-2, C-7 and C-9 positions. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Quinolines and derivatives |
|---|
| Sub Class | Pyrroloquinolines |
|---|
| Direct Parent | Pyrroloquinoline quinones |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyrroloquinoline quinone
- Quinoline-4-carboxylic acid
- Quinoline-2-carboxylic acid
- Indole or derivatives
- Pyridine carboxylic acid
- Pyridine carboxylic acid or derivatives
- Tricarboxylic acid or derivatives
- O-quinone
- Pyrrole-2-carboxylic acid
- Pyrrole-2-carboxylic acid or derivatives
- Aryl ketone
- Quinone
- Pyridine
- Substituted pyrrole
- Heteroaromatic compound
- Pyrrole
- Vinylogous amide
- Ketone
- Azacycle
- Carboxylic acid derivative
- Carboxylic acid
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Organooxygen compound
- Organonitrogen compound
- Organic oxygen compound
- Organic nitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -3 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0rkl-1293000000-f89d3e9669ea62420001 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-00di-5001920000-ffb897aa343201141554 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03dr-0059000000-fe32e3dcdbb122899a9b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01p9-0093000000-0057ba82bb1928e2b729 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00kf-0090000000-7a80db950a195ffc246a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-01ti-0089000000-3940ecb0162179ef62fc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00kr-0092000000-47c12a164c99ff5da573 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00ku-0090000000-968a80882216e4efda23 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0009000000-84aa1fbc000600884df5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-02ai-0059000000-1b2548c8675bc744991a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-014l-0094000000-fd415930659367c11a26 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0090000000-984e546238980fbb8d35 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-0090000000-899cb3d0e242698d7cf3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-0090000000-899cb3d0e242698d7cf3 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Kasahara, T., Kato, T. (2003). "Nutritional biochemistry: A new redox-cofactor vitamin for mammals." Nature 422:832. Pubmed: 12712191
- Paz, M. A., Fluckiger, R., Torrelio, B. M., Gallop, P. M. (1989). "Methoxatin (PQQ), coenzyme for copper-dependent amine and mixed-function oxidation in mammalian tissues." Connect Tissue Res 20:251-257. Pubmed: 2558842
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
- Winder, C. L., Dunn, W. B., Schuler, S., Broadhurst, D., Jarvis, R., Stephens, G. M., Goodacre, R. (2008). "Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites." Anal Chem 80:2939-2948. Pubmed: 18331064
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|