| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-08-09 09:16:14 -0600 |
|---|
| Update Date | 2015-09-13 12:56:16 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
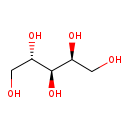
| Name: | L-Arabitol |
|---|
| Description | L-Arabitol or arabinitol is a sugar alcohol. It can be formed by the reduction of either arabinose or lyxose. |
|---|
| Structure | |
|---|
| Synonyms: | - 1,2,3,4,5-Pentahydroxypentane
- 1,2,3,4,5-Pentanepentol
- Adonite Adonitol
- Arabinitol
- Arabitol
- L(-)-Arabitol
- L-(+)-Arabitol
- L-(-)-Arabinitol
- L-(-)-Arabitol
- L-Arabinitol
- L-Arabinol
- L-Lyxitol
|
|---|
| Chemical Formula: | C5H12O5 |
|---|
| Weight: | Average: 152.1458
Monoisotopic: 152.068473494 |
|---|
| InChI Key: | HEBKCHPVOIAQTA-IMJSIDKUSA-N |
|---|
| InChI: | InChI=1S/C5H12O5/c6-1-3(8)5(10)4(9)2-7/h3-10H,1-2H2/t3-,4-/m0/s1 |
|---|
| CAS number: | 7643-75-6 |
|---|
| IUPAC Name: | (2S,4S)-pentane-1,2,3,4,5-pentol |
|---|
| Traditional IUPAC Name: | arabinitol |
|---|
| SMILES: | OC[C@H](O)C(O)[C@@H](O)CO |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as sugar alcohols. These are hydrogenated forms of carbohydrate in which the carbonyl group (aldehyde or ketone, reducing sugar) has been reduced to a primary or secondary hydroxyl group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Sugar alcohols |
|---|
| Alternative Parents | |
|---|
| Substituents | - Sugar alcohol
- Monosaccharide
- Secondary alcohol
- Polyol
- Hydrocarbon derivative
- Primary alcohol
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | 0 |
|---|
| Melting point: | 101 - 104 °C |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (5 TMS) | splash10-0uxs-0930000000-6ef7d2f1643b576f6fbf | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0uxs-0930000000-6ef7d2f1643b576f6fbf | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-03dl-9100000000-51c1d4635c9c103902c4 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (5 TMS) - 70eV, Positive | splash10-0a4i-6033900000-a5ac27dc62580d112ccd | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-00di-6900000000-0ddb428b63c2d2c275b1 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0abl-9000000000-01b0114148e5ae6b5689 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0a4i-9000000000-d30546aff0a9d5533991 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9000000000-90a5e5f7f537d92a3b01 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0kg9-9300000000-ecec51f40684a2bda88c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0ab9-9000000000-3a20dd45912bfcec959f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-0uki-9600000000-1f9f66e6ca234852798b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-3bde5af4fd67b6ab1193 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-066r-9000000000-e3badf09bef4f78c4044 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-066r-9000000000-4f277ce5896e451f35d6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-1900000000-4f856d537cc8252ecc47 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-9300000000-403bacad5117a30582df | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03dl-9000000000-eb879b27e7b182cfe7c0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udu-9400000000-3db6454ab3e2315aa744 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0btl-9200000000-b84c400427c7e44eb4dd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9000000000-c37624fb31a8cb33b163 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0zmr-9500000000-1887747b16d168b09609 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-9000000000-83f8fb1c321a951b0e7e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9000000000-ad178d7c96ba63131c18 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-4900000000-d02566d106f64f56452d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01ox-9000000000-94760e1883352d4864b5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0007-9000000000-a22bb5556fa857db4c5c | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H,1H] 2D NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Onkenhout W, Groener JE, Verhoeven NM, Yin C, Laan LA: L-Arabinosuria: a new defect in human pentose metabolism. Mol Genet Metab. 2002 Sep-Oct;77(1-2):80-5. Pubmed: 12359133
- TOUSTER O, HARWELL SO: The isolation of L-arabitol from pentosuric urine. J Biol Chem. 1958 Feb;230(2):1031-41. Pubmed: 13525419
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Download (PDF) |
|---|
| Links |
|---|
| External Links: | |
|---|