Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
gamma-Glutamyl-gamma-butyraldehyde (M2MDB001737)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-07-30 14:55:21 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 17:21:11 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | gamma-Glutamyl-gamma-butyraldehyde | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Gamma-glutamyl-gamma-butyraldehyde is a member of the chemical class known as Alpha Amino Acids and Derivatives. These are amino acids in which the amino group is attached to the carbon atom immediately adjacent to the carboxylate group (alpha carbon).Gamma-glutamyl-gamma-butyraldehyde is invovled in Arginine and proline metabolism. (KEGG) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
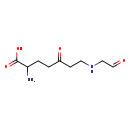
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C9H16N2O4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 216.2343 Monoisotopic: 216.11100701 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | SSNXXAXHQSXHKM-UHFFFAOYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C9H16N2O4/c10-8(9(14)15)2-1-7(13)3-4-11-5-6-12/h6,8,11H,1-5,10H2,(H,14,15) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | 2-amino-5-oxo-7-[(2-oxoethyl)amino]heptanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | 2-amino-5-oxo-7-[(2-oxoethyl)amino]heptanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | NC(CCC(=O)CCNCC=O)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as alpha amino acids. These are amino acids in which the amino group is attached to the carbon atom immediately adjacent to the carboxylate group (alpha carbon). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Amino acids, peptides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Alpha amino acids | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | gamma-Glutamyl-gamma-butyraldehyde + Water + NADP <> 4-(Glutamylamino) butanoate +2 Hydrogen ion + NADPH gamma-Glutamyl-L-putrescine + Water + Oxygen > gamma-Glutamyl-gamma-butyraldehyde + Hydrogen peroxide + Ammonium gamma-Glutamyl-L-putrescine + Water + Oxygen <> gamma-Glutamyl-gamma-butyraldehyde + Ammonia + Hydrogen peroxide gamma-Glutamyl-gamma-butyraldehyde + NAD + Water <> 4-(Glutamylamino) butanoate + NADH + Hydrogen ion gamma-Glutamyl-gamma-butyraldehyde + NADP + Water <> 4-(Glutamylamino) butanoate + NADPH + Hydrogen ion NAD(P)<sup>+</sup> + gamma-Glutamyl-gamma-butyraldehyde + Water > 4-(Glutamylamino) butanoate + NAD(P)H + Hydrogen ion gamma-Glutamyl-L-putrescine + Oxygen + Hydrogen ion > gamma-Glutamyl-gamma-butyraldehyde + Hydrogen peroxide + Ammonium gamma-Glutamyl-gamma-butyraldehyde + NADP + Water > gamma-glutamyl-gamma-aminobutyrate + NADPH +2 Hydrogen ion gamma-Glutamyl-L-putrescine + Water + Oxygen <> gamma-Glutamyl-gamma-butyraldehyde + Ammonia + Hydrogen peroxide gamma-Glutamyl-gamma-butyraldehyde + Water + NADP <>4 4-(Glutamylamino) butanoate +2 Hydrogen ion + NADPH | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in oxidoreductase activity
- Specific function:
- Involved in the breakdown of putrescine. Was previously shown to have a weak but measurable ALDH enzyme activity that prefers NADP over NAD as coenzyme
- Gene Name:
- puuC
- Uniprot ID:
- P23883
- Molecular weight:
- 53418
Reactions
| Gamma-glutamyl-gamma-aminobutyraldehyde + NAD(+) + H(2)O = gamma-glutamyl-gamma-aminobutyrate + NADH. |
| An aldehyde + NAD(P)(+) + H(2)O = a carboxylate + NAD(P)H. |
- General function:
- Involved in oxidoreductase activity
- Specific function:
- Involved in the breakdown of putrescine via the oxidation of L-glutamylputrescine
- Gene Name:
- puuB
- Uniprot ID:
- P37906
- Molecular weight:
- 47169
Reactions
| Gamma-glutamylputrescine + H(2)O + O(2) = Gamma-glutamyl-gamma-aminobutyraldehyde + NH(3) + H(2)O(2). |