Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
Quinate (M2MDB001685)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-07-30 14:55:12 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 17:21:04 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Quinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Quinic acid is a cyclitol, a cyclic polyol. It is a crystalline acid obtained from cinchona bark, coffee beans, and other plant products and made synthetically by hydrolysis of chlorogenic acid. Quinic acid is also implicated in the perceived acidity of coffee. It is a constituent of the tara tannins.; This acid is a versatile chiral starting material for the synthesis of new pharmaceuticals. A new medicament for the treatment of influenza A and B strains called Tamiflu has been successfully developed and launched into the market. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
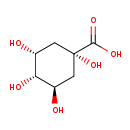
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C7H12O6 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 192.1666 Monoisotopic: 192.063388116 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | AAWZDTNXLSGCEK-WYWMIBKRSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C7H12O6/c8-3-1-7(13,6(11)12)2-4(9)5(3)10/h3-5,8-10,13H,1-2H2,(H,11,12)/t3-,4-,5-,7+/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 17230-85-2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexane-1-carboxylic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | (-)-quinic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | O[C@@H]1C[C@@](O)(C[C@@H](O)[C@H]1O)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as quinic acids and derivatives. Quinic acids and derivatives are compounds containing a quinic acid moiety (or a derivative thereof), which is a cyclitol made up of a cyclohexane ring that bears four hydroxyl groups at positions 1,3.4, and 5, as well as a carboxylic acid at position 1. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic oxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Alcohols and polyols | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Quinic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic homomonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 168 °C, 441 K, 334 °F | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | NAD + Quinate <> 3-Dehydroquinate +2 Hydrogen ion + NADH Quinate + NAD <> 3-Dehydroquinate + NADH + Hydrogen ion Quinate + NADP <> 3-Dehydroquinate + NADPH + Hydrogen ion NAD(P)<sup>+</sup> + Quinate NAD(P)H + 3-Dehydroquinate + Hydrogen ion Quinate + NAD(P)(+) > 3-Dehydroquinate + NAD(P)H Quinate + NAD + NADP + Shikimic acid <> 3-Dehydroquinate + NADH + NADPH + Hydrogen ion + 3-Dehydro-shikimate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in nucleotide binding
- Specific function:
- The physiological substrate is not known
- Gene Name:
- ydiB
- Uniprot ID:
- P0A6D5
- Molecular weight:
- 31228
Reactions
| L-quinate + NAD(P)(+) = 3-dehydroquinate + NAD(P)H. |
| Shikimate + NAD(P)(+) = 3-dehydroshikimate + NAD(P)H. |
Transporters
- General function:
- Involved in transporter activity
- Specific function:
- Non-specific porin
- Gene Name:
- ompN
- Uniprot ID:
- P77747
- Molecular weight:
- 41220
- General function:
- Involved in transporter activity
- Specific function:
- Uptake of inorganic phosphate, phosphorylated compounds, and some other negatively charged solutes
- Gene Name:
- phoE
- Uniprot ID:
- P02932
- Molecular weight:
- 38922
- General function:
- Involved in transporter activity
- Specific function:
- OmpF is a porin that forms passive diffusion pores which allow small molecular weight hydrophilic materials across the outer membrane. It is also a receptor for the bacteriophage T2
- Gene Name:
- ompF
- Uniprot ID:
- P02931
- Molecular weight:
- 39333
- General function:
- Involved in transporter activity
- Specific function:
- Forms passive diffusion pores which allow small molecular weight hydrophilic materials across the outer membrane
- Gene Name:
- ompC
- Uniprot ID:
- P06996
- Molecular weight:
- 40368