| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-07-30 14:55:07 -0600 |
|---|
| Update Date | 2015-06-03 17:21:00 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | N-Methyltryptophan |
|---|
| Description | N-methyltryptophan is a member of the chemical class known as Alpha Amino Acids and Derivatives. These are amino acids in which the amino group is attached to the carbon atom immediately adjacent to the carboxylate group. N-Methyltryptophan is catabolized by MTOX. N-Methyltryptophan oxidase (MTOX) is a flavoenzyme that catalyzes the oxidative demethylation of N-methyl-L-tryptophan and other N-methyl amino acids, including sarcosine, which is a poor substrate. (PMID 11170472) MTOX is converted to a 2-electron reduced form upon anaerobic reaction with N-methyl-L-tryptophan, sarcosine, or the carbinolamine formed with L-tryptophan and formaldehyde. (PMID 11170473) |
|---|
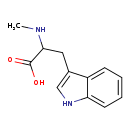
| Structure | |
|---|
| Synonyms: | - (2S)-3-(1H-indol-3-yl)-2-(methylamino)propanoate
- (2S)-3-(1H-indol-3-yl)-2-(methylamino)propanoic acid
- Abrine
- L-2-Methyltryptophan
- L-Abrine
- N-Methyl-L-tryptophan
- N-Methyl-L-tryptophane
|
|---|
| Chemical Formula: | C12H14N2O2 |
|---|
| Weight: | Average: 218.2518
Monoisotopic: 218.105527702 |
|---|
| InChI Key: | CZCIKBSVHDNIDH-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/C12H14N2O2/c1-13-11(12(15)16)6-8-7-14-10-5-3-2-4-9(8)10/h2-5,7,11,13-14H,6H2,1H3,(H,15,16) |
|---|
| CAS number: | 526-31-8 |
|---|
| IUPAC Name: | 3-(1H-indol-3-yl)-2-(methylamino)propanoic acid |
|---|
| Traditional IUPAC Name: | 3-(1H-indol-3-yl)-2-(methylamino)propanoic acid |
|---|
| SMILES: | CNC(CC1=CNC2=CC=CC=C12)C(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as alpha amino acids. These are amino acids in which the amino group is attached to the carbon atom immediately adjacent to the carboxylate group (alpha carbon). |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Alpha amino acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-amino acid
- 3-alkylindole
- Indole
- Indole or derivatives
- Aralkylamine
- Substituted pyrrole
- Benzenoid
- Heteroaromatic compound
- Pyrrole
- Amino acid
- Azacycle
- Organoheterocyclic compound
- Carboxylic acid
- Secondary aliphatic amine
- Secondary amine
- Monocarboxylic acid or derivatives
- Hydrocarbon derivative
- Organonitrogen compound
- Organooxygen compound
- Organopnictogen compound
- Organic oxygen compound
- Organic nitrogen compound
- Organic oxide
- Carbonyl group
- Amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | 0 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | |
|---|
| References |
|---|
| References: | - van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
- Winder, C. L., Dunn, W. B., Schuler, S., Broadhurst, D., Jarvis, R., Stephens, G. M., Goodacre, R. (2008). "Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites." Anal Chem 80:2939-2948. Pubmed: 18331064
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|