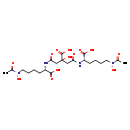| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-07-30 14:54:48 -0600 |
|---|
| Update Date | 2015-06-03 17:20:50 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | Aerobactin |
|---|
| Description | Aerobactin is a iron chelating agent (siderophore) found in bacteria such as E. coli, allowing bacteria to transport and sequester iron. It is a virulence factor enabling E. coli to sequester iron in iron-poor environments such as host urinary tracts. (Wikipedia, ChEBI) Aerobactin can be made in E. coli through the degradation of lysine by the enzyme aerobactin synthase (EC 6.3.2.27). (Wikipedia, KEGG) |
|---|
| Structure | |
|---|
| Synonyms: | - (8S,16S)-3,12,21-trihydroxy-2,10,14,22-tetraoxo-3,9,15,21-tetraazatricosane-8,12,16-tricarboxylate
- (8S,16S)-3,12,21-trihydroxy-2,10,14,22-tetraoxo-3,9,15,21-tetraazatricosane-8,12,16-tricarboxylic acid
- Aerobactin
- Ferric-aerobactin
- N2,N2'-(3-carboxy-3-hydroxypentanedioyl)bis(N6-acetyl-N6-hydroxy-L-lysine)
|
|---|
| Chemical Formula: | C22H36N4O13 |
|---|
| Weight: | Average: 564.5402
Monoisotopic: 564.227887258 |
|---|
| InChI Key: | KDHHWXGBNUCREU-HOTGVXAUSA-N |
|---|
| InChI: | InChI=1S/C22H36N4O13/c1-13(27)25(38)9-5-3-7-15(19(31)32)23-17(29)11-22(37,21(35)36)12-18(30)24-16(20(33)34)8-4-6-10-26(39)14(2)28/h15-16,37-39H,3-12H2,1-2H3,(H,23,29)(H,24,30)(H,31,32)(H,33,34)(H,35,36)/t15-,16-/m0/s1 |
|---|
| CAS number: | 26198-65-2 |
|---|
| IUPAC Name: | (2S)-2-[3-carboxy-3-({[(1S)-1-carboxy-5-(N-hydroxyacetamido)pentyl]carbamoyl}methyl)-3-hydroxypropanamido]-6-(N-hydroxyacetamido)hexanoic acid |
|---|
| Traditional IUPAC Name: | aerobactin |
|---|
| SMILES: | CC(=O)N(O)CCCC[C@H](NC(=O)CC(O)(CC(=O)N[C@@H](CCCCN(O)C(C)=O)C(O)=O)C(O)=O)C(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as n-acyl-alpha amino acids. N-acyl-alpha amino acids are compounds containing an alpha amino acid which bears an acyl group at its terminal nitrogen atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | N-acyl-alpha amino acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - N-acyl-alpha-amino acid
- Tricarboxylic acid or derivatives
- Alpha-hydroxy acid
- Hydroxy acid
- Tertiary alcohol
- Acetohydroxamic acid
- Acetamide
- Hydroxamic acid
- Carboximidic acid
- Carboximidic acid derivative
- Carboxylic acid
- Organic 1,3-dipolar compound
- Propargyl-type 1,3-dipolar organic compound
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Organic nitrogen compound
- Organonitrogen compound
- Organooxygen compound
- Hydrocarbon derivative
- Carbonyl group
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -3 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00kf-9114360000-aae8c90dd540d7190ae4 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-0006-6004191000-a3d9063b76c1b9fcce3c | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("Aerobactin,1TMS,#1" TMS) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0111190000-6cceaaf6740bbfb89d96 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-052b-0791880000-fb56d3b8852220ec731a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-2962330000-5df621761fc9c7904251 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-01b9-2024090000-7b805b2f239cf5b4ee97 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0v4j-2192280000-21815bd606980decb1a5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0ufr-2291010000-4d44fb96363a2d143039 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-05fr-0000290000-4709b636547109837686 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-0321390000-5cfd6c97627637f03efe | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-053u-2910300000-c2c77f243e63232cf5f2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0fka-2000190000-14f7c8f7e438026bb6c8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-1011290000-aee3ea2f25dfea9dc2d0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0f89-2291100000-221052676a4a2ac8f391 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Keller R, Pedroso MZ, Ritchmann R, Silva RM: Occurrence of virulence-associated properties in Enterobacter cloacae. Infect Immun. 1998 Feb;66(2):645-9. Pubmed: 9453621
- Opal SM, Cross AS, Gemski P, Lyhte LW: Aerobactin and alpha-hemolysin as virulence determinants in Escherichia coli isolated from human blood, urine, and stool. J Infect Dis. 1990 Apr;161(4):794-6. Pubmed: 2181035
- Schlager TA, Whittam TS, Hendley JO, Wilson RA, Bhang J, Grady R, Stapleton A: Expression of virulence factors among Escherichia coli isolated from the periurethra and urine of children with neurogenic bladder on intermittent catheterization. Pediatr Infect Dis J. 2000 Jan;19(1):37-41. Pubmed: 10643848
- Sharma S, Harjai K, Mittal R: Enhanced siderophore production and mouse kidney pathogenicity in Escherichia coli grown in urine. J Med Microbiol. 1991 Dec;35(6):325-9. Pubmed: 1836502
- Vazquez F, Mendoza MC, Viejo G, Mendez FJ: Survey of Escherichia coli septicemia over a six-year period. Eur J Clin Microbiol Infect Dis. 1992 Feb;11(2):110-7. Pubmed: 1396724
- Williams PH, Carbonetti NH: Iron, siderophores, and the pursuit of virulence: independence of the aerobactin and enterochelin iron uptake systems in Escherichia coli. Infect Immun. 1986 Mar;51(3):942-7. Pubmed: 2936686
- Yamamoto S, Shinoda S: [Iron uptake mechanisms of pathogenic bacteria]. Nihon Saikingaku Zasshi. 1996 Feb;51(2):523-47. Pubmed: 8752377
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|