Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
7-Deaza-7-carboxyguanine (M2MDB001592)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-07-30 14:54:47 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 17:20:49 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | 7-Deaza-7-carboxyguanine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | 7-deaza-7-carboxyguanine is a member of the chemical class known as Pyrrolopyrimidines. These are compounds containing a pyrrolopyrimidine moiety, which consists of a pyrrole ring fused to a pyrimidine. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
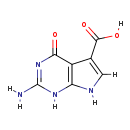
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C7H6N4O3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 194.1475 Monoisotopic: 194.043990078 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | XIUIRSLBMMTDSK-UHFFFAOYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C7H6N4O3/c8-7-10-4-3(5(12)11-7)2(1-9-4)6(13)14/h1H,(H,13,14)(H4,8,9,10,11,12) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | 2-amino-4-oxo-1H,4H,7H-pyrrolo[2,3-d]pyrimidine-5-carboxylic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | 2-amino-4-oxo-1H,7H-pyrrolo[2,3-d]pyrimidine-5-carboxylic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | [H]OC(=O)C1=C([H])N([H])C2=C1C(=O)N=C(N([H])[H])N2[H] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as pyrrolo[2,3-d]pyrimidines. These are aromatic heteropolycyclic compounds containing a pyrrolo[2,3-d]pyrimidine ring system, which is an pyrrolopyrimidine isomers having the 3 ring nitrogen atoms at the 1-, 5-, and 7-positions. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organoheterocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Pyrrolopyrimidines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Pyrrolo[2,3-d]pyrimidines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Pyrrolo[2,3-d]pyrimidines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosine triphosphate + 7-Deaza-7-carboxyguanine + Ammonium > ADP + Hydrogen ion + Water + Phosphate + 7-Cyano-7-carbaguanine 6-Carboxy-5,6,7,8-tetrahydropterin <> 7-Carboxy-7-carbaguanine + Ammonia + 7-Deaza-7-carboxyguanine 7-Deaza-7-carboxyguanine + Ammonia + Adenosine triphosphate > 7-Cyano-7-carbaguanine + ADP + Inorganic phosphate + Water 6-Carboxy-5,6,7,8-tetrahydropterin > 7-Deaza-7-carboxyguanine + Ammonia 7-Deaza-7-carboxyguanine + Ammonia + Adenosine triphosphate <> 7-Cyano-7-carbaguanine + ADP + Phosphate + Water 7-Deaza-7-carboxyguanine + Adenosine triphosphate + Ammonium > Water + Phosphate + Adenosine diphosphate + Hydrogen ion + 7-Cyano-7-carbaguanine + ADP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in argininosuccinate synthase activity
- Specific function:
- Catalyzes the ATP-dependent conversion of 7-carboxy-7- deazaguanine (CDG) to 7-cyano-7-deazaguanine (preQ(0))
- Gene Name:
- queC
- Uniprot ID:
- P77756
- Molecular weight:
- 25514
Reactions
| 7-carboxy-7-carbaguanine + NH(3) + ATP = 7-cyano-7-carbaguanine + ADP + phosphate + H(2)O. |
- General function:
- Not Available
- Specific function:
- Not Available
- Gene Name:
- queE
- Uniprot ID:
- P64554
- Molecular weight:
- Not Available