| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 14:50:59 -0600 |
|---|
| Update Date | 2015-06-03 17:20:18 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | Homoserine lactone |
|---|
| Description | N-Acyl homoserine lactones (AHLs or N-AHLs) are a class of signaling molecules involved in bacterial quorum sensing. Quorum sensing is a method of communication between bacteria that enables the coordination of group based behavior based on population density. They signal changes in gene expression, such as switching between the flagella gene and the gene for pili for the development of a biofilm.One example of the involvement of AHLs in quorum sensing is in the regulation of the bioluminescent protein luciferase in the luminescent bacteria Vibrio fischeri. Similar pathways occur in other luminescent bacteria. In Vibrio fischeri, when the concentration of AHLs exceeds a certain limit, the transcription of the operon luxAB begins. Transcription of the operon results in luminescence due to the expression of LuxA and LuxB, which form a protein known as luciferase. This is an important feature of quorum sensing, as it makes little sense for one cell to waste the energy producing light, as the resulting light will be so faint it will be more or less undetectable. Instead, once the bacterial population has reached a specific size, only then does light production commence. |
|---|
| Structure | |
|---|
| Synonyms: | - α-amino-γ-butyrolactone
- 2(3H)-furanone, 3-aminodihydro-
- 2-Aminobutan-4-olide
- 3-Aminodihydro-2(3H)-Furanone
- a-amino-g-Butyrolactone
- Alpha-Amino-gamma-butyrolactone
- Hsl
- N-Butanoyl-lhomoserine lactone
- N-Butyryl-DL-homoserine lactone
- N-Butyryl-L-homoserine lactone
- N-Butyrylhomoserine lactone
- α-amino-γ-Butyrolactone
|
|---|
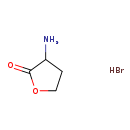
| Chemical Formula: | C4H8BrNO2 |
|---|
| Weight: | Average: 182.016
Monoisotopic: 180.973841152 |
|---|
| InChI Key: | MKLNTBLOABOJFZ-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/C4H7NO2.BrH/c5-3-1-2-7-4(3)6;/h3H,1-2,5H2;1H |
|---|
| CAS number: | 1192-20-7 |
|---|
| IUPAC Name: | 3-aminooxolan-2-one hydrobromide |
|---|
| Traditional IUPAC Name: | HSLs hydrobromide |
|---|
| SMILES: | Br.NC1CCOC1=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as alpha amino acid esters. These are ester derivatives of alpha amino acids. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Alpha amino acid esters |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-amino acid ester
- Gamma butyrolactone
- Tetrahydrofuran
- Carboxylic acid ester
- Lactone
- Monocarboxylic acid or derivatives
- Organoheterocyclic compound
- Oxacycle
- Hydrocarbon derivative
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Primary aliphatic amine
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Carbonyl group
- Organic nitrogen compound
- Hydrobromide
- Amine
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | 1 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | |
|---|
| References |
|---|
| References: | - Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|