Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
Fructoselysine-6-phosphate (M2MDB001363)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-05-31 14:50:34 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 17:20:18 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Fructoselysine-6-phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Fructoselysine-6-phosphate is a member of the chemical class known as Alpha Amino Acids and Derivatives. These are amino acids in which the amino group is attached to the carbon atom immediately adjacent to the carboxylate group (alpha carbon).Fructoselysine-6-phosphate is invovled in Fructoselysine degradation. Fructoselysine-6-phosphate is involved in the metabolism of fructation product. The metabolism of the glycation product fructose-epsilon-lysine in Escherichia coli involves its ATP-dependent phosphorylation by a specific kinase (FrlD), followed by the conversion of fructoselysine 6-phosphate into glucose 6-phosphate and lysine by fructoselysine-6-phosphate deglycase (FrlB), which is distantly related to the isomerase domain of glucosamine-6-phosphate synthase. (PMID 16153181) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
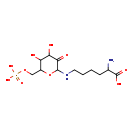
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C12H23N2O10P | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 386.2922 Monoisotopic: 386.109031478 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | BZFCRRVQZCIOBA-UHFFFAOYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C12H23N2O10P/c13-6(12(18)19)3-1-2-4-14-11-10(17)9(16)8(15)7(24-11)5-23-25(20,21)22/h6-9,11,14-16H,1-5,13H2,(H,18,19)(H2,20,21,22) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | 2-amino-6-({4,5-dihydroxy-3-oxo-6-[(phosphonooxy)methyl]oxan-2-yl}amino)hexanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | 2-amino-6-({4,5-dihydroxy-3-oxo-6-[(phosphonooxy)methyl]oxan-2-yl}amino)hexanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | NC(CCCCNC1OC(COP(O)(O)=O)C(O)C(O)C1=O)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as alpha amino acids. These are amino acids in which the amino group is attached to the carbon atom immediately adjacent to the carboxylate group (alpha carbon). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Amino acids, peptides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Alpha amino acids | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic heteromonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Fructoselysine-6-phosphate + Water <> Glucose 6-phosphate + L-Lysine Adenosine triphosphate + Fructoselysine > ADP + Fructoselysine-6-phosphate + Hydrogen ion Fructoselysine-6-phosphate + Water > Glucose 6-phosphate + L-Lysine Adenosine triphosphate + Fructoselysine > ADP + Fructoselysine-6-phosphate 1-[(5-Amino-5-carboxypentyl)amino]-1-deoxyfructose + Adenosine triphosphate > Fructoselysine-6-phosphate + ADP + Hydrogen ion Fructoselysine-6-phosphate + Water <> β-D-glucose 6-phosphate + L-Lysine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in carbohydrate binding
- Specific function:
- Catalyzes the conversion of fructoselysine 6-phosphate to glucose 6-phosphate and lysine
- Gene Name:
- frlB
- Uniprot ID:
- P0AC00
- Molecular weight:
- 38569
Reactions
| Fructoselysine 6-phosphate + H(2)O = glucose 6-phosphate + L-lysine. |
- General function:
- Involved in phosphotransferase activity, alcohol group as acceptor
- Specific function:
- Phosphorylates fructoselysine to yield fructoselysine 6- phosphate
- Gene Name:
- frlD
- Uniprot ID:
- P45543
- Molecular weight:
- 28332
Reactions
| ATP + fructoselysine = ADP + fructoselysine 6-phosphate. |