Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
Sirohydrochlorin (M2MDB001041)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-05-31 14:32:32 -0600 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 17:19:30 -0600 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Sirohydrochlorin | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Sirohydrochlorin is a member of the chemical class known as Precorrins. These are intermediates formed by methylation at one or more of the four rings prior to the formation of the macrocyclic corrin ring. typhimurium, precorrin-2 is a precursor of both siroheme and B12. (PMID 8955319) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
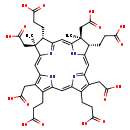
| Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C42H46N4O16 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 862.8318 Monoisotopic: 862.290881444 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | KWIZRXMMFRBUML-AHGFGAHVSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C42H46N4O16/c1-41(17-39(59)60)23(5-9-35(51)52)29-14-27-21(11-37(55)56)19(3-7-33(47)48)25(43-27)13-26-20(4-8-34(49)50)22(12-38(57)58)28(44-26)15-31-42(2,18-40(61)62)24(6-10-36(53)54)30(46-31)16-32(41)45-29/h13-16,23-24,44-45H,3-12,17-18H2,1-2H3,(H,47,48)(H,49,50)(H,51,52)(H,53,54)(H,55,56)(H,57,58)(H,59,60)(H,61,62)/b25-13-,29-14-,31-15-,32-16-/t23-,24-,41+,42+/m1/s1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 65207-12-7 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | 3-[(4S,5S,9S,10S)-9,15,19-tris(2-carboxyethyl)-5,10,14,20-tetrakis(carboxymethyl)-5,10-dimethyl-21,22,23,24-tetraazapentacyclo[16.2.1.1³,⁶.1⁸,¹¹.1¹³,¹⁶]tetracosa-1(21),2,6,8(23),11,13,15,17,19-nonaen-4-yl]propanoic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | sirohydrochlorin | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | C[C@]1(CC(O)=O)[C@H](CCC(O)=O)\C2=C\C3=N\C(=C/C4=C(CCC(O)=O)C(CC(O)=O)=C(N4)\C=C4/N=C(/C=C1\N2)[C@@H](CCC(O)=O)[C@]4(C)CC(O)=O)\C(CCC(O)=O)=C3CC(O)=O | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as tetrapyrroles and derivatives. These are polycyclic aromatic compounds containing four pyrrole rings joined by one-carbon units linking position 2 of one pyrrole ring to position 5 of the next. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organoheterocyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Tetrapyrroles and derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Tetrapyrroles and derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -8 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Membrane | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Iron + Sirohydrochlorin >3 Hydrogen ion + Siroheme Precorrin 2 + NAD > Hydrogen ion + NADH + Sirohydrochlorin Siroheme + 2 Hydrogen ion <> Fe2+ + Sirohydrochlorin Precorrin 2 + NAD <> Sirohydrochlorin + NADH + Hydrogen ion Sirohydrochlorin + Iron <> Hydrogen ion + Siroheme Precorrin-2 + NAD > Sirohydrochlorin + NADH Siroheme + 2 Hydrogen ion > Sirohydrochlorin + Iron Precorrin-2 + NAD > NADH + Hydrogen ion + Sirohydrochlorin Sirohydrochlorin + Iron >2 Hydrogen ion + Siroheme Precorrin 2 + NAD <> Sirohydrochlorin + NADH +2 Hydrogen ion | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in methyltransferase activity
- Specific function:
- Multifunctional enzyme that catalyzes the SAM-dependent methylation of uroporphyrinogen III at position C-2 and C-7 to form precorrin-2 and then position C-12 or C-18 to form trimethylpyrrocorphin 2. It also catalyzes the conversion of precorrin-2 into siroheme. This reaction consists of the NAD- dependent oxidation of precorrin-2 into sirohydrochlorin and its subsequent ferrochelation into siroheme
- Gene Name:
- cysG
- Uniprot ID:
- P0AEA8
- Molecular weight:
- 49951
Reactions
| S-adenosyl-L-methionine + uroporphyrinogen III = S-adenosyl-L-homocysteine + precorrin-1. |
| S-adenosyl-L-methionine + precorrin-1 = S-adenosyl-L-homocysteine + precorrin-2. |
| Precorrin-2 + NAD(+) = sirohydrochlorin + NADH. |
| Siroheme + 2 H(+) = sirohydrochlorin + Fe(2+). |