| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 14:31:20 -0600 |
|---|
| Update Date | 2015-06-03 17:19:27 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
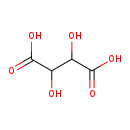
| Name: | Meso-Tartaric acid |
|---|
| Description | Meso-tartaric acid is a member of the chemical class known as Dicarboxylic Acids and Derivatives. These are organic compounds containing exactly two carboxylic acid groups. |
|---|
| Structure | |
|---|
| Synonyms: | - (2R,3S)-2,3-dihydroxybutanedioate
- (2R,3S)-2,3-dihydroxybutanedioic acid
- (2R,3S)-2,3-dihydroxysuccinate
- (2R,3S)-2,3-dihydroxysuccinic acid
- (2R,3S)-rel-2,3-dihydroxybutanedioate
- (2R,3S)-rel-2,3-dihydroxybutanedioic acid
- (2R,3S)-tartarate
- (2R,3S)-tartaric acid
- (R*,S*)-2,3-dihydroxybutanedioate
- (R*,S*)-2,3-dihydroxybutanedioic acid
- 1,4-BUTANEDIOate,2,3-dihydroxy (tartarate)
- 1,4-BUTANEDIOIC ACID,2,3-DIHYDROXY (TARTARIC ACID)
- 2,3-Dihydroxy-(R*,S*)-Butanedioate
- 2,3-Dihydroxy-(R*,S*)-Butanedioic acid
- 2,3-Dihydroxy-Butanedioate
- 2,3-Dihydroxy-Butanedioic acid
- 2,3-Dihydroxysuccinate
- 2,3-Dihydroxysuccinic acid
- meso-tartaric acid
- Erythrarate
- Erythraric acid
- Internally compensate tartarate
- Internally compensate tartaric acid
- Internally compensated tartarate
- Internally compensated tartaric acid
- Internally compensic acid tartaric acid
- M-Tartarate
- M-Tartaric acid
- Meso-Tartarate
- Meso-tartarate monohydrate
- Meso-tartaric acid monohydrate
- Meso-tartaric acid monohydric acid
- Meso-tartrate
- Meso-tartric acid
- Mesotartarate
- Mesotartaric acid
- Mesoweinsaeure
- S,R Meso-tartarate
- S,R Meso-tartaric acid
- SRT
- Unresolvable tartarate
- Unresolvable tartaric acid
|
|---|
| Chemical Formula: | C4H6O6 |
|---|
| Weight: | Average: 150.0868
Monoisotopic: 150.016437924 |
|---|
| InChI Key: | FEWJPZIEWOKRBE-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/C4H6O6/c5-1(3(7)8)2(6)4(9)10/h1-2,5-6H,(H,7,8)(H,9,10) |
|---|
| CAS number: | 147-73-9 |
|---|
| IUPAC Name: | 2,3-dihydroxybutanedioic acid |
|---|
| Traditional IUPAC Name: | (.+-.)-tartaric acid |
|---|
| SMILES: | OC(C(O)C(O)=O)C(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as sugar acids and derivatives. Sugar acids and derivatives are compounds containing a saccharide unit which bears a carboxylic acid group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Sugar acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Sugar acid
- Short-chain hydroxy acid
- Beta-hydroxy acid
- Fatty acid
- Monosaccharide
- Hydroxy acid
- Dicarboxylic acid or derivatives
- Alpha-hydroxy acid
- Secondary alcohol
- 1,2-diol
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxide
- Hydrocarbon derivative
- Carbonyl group
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -2 |
|---|
| Melting point: | 147 °C |
|---|
| Experimental Properties: | | Property | Value | Source |
|---|
| Water Solubility: | 560 mg/mL at 20 oC [YALKOWSKY,SH & DANNENFELSER,RM (1992)] | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | - Glyoxylate and dicarboxylate metabolism ec00630
|
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0002-0920000000-473bf90ce56c0c0592a0 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0002-0920000000-473bf90ce56c0c0592a0 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0002-0920000000-473bf90ce56c0c0592a0 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0002-0920000000-473bf90ce56c0c0592a0 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0096-9200000000-df0ce9fba754b55674b1 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-00di-8029100000-283552abf9161a349136 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-0002-5900000000-4c66eac9e5850c5fcbe4 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - , negative | splash10-0072-9600000000-17c4d1ba2b96cd7ef84c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - , negative | splash10-0072-9600000000-f9fb12efb15f5ed29bce | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0072-9600000000-4115648caec12719bc40 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-00du-9000000000-76ecadb353351b98d37d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0f89-1900000000-81029c7f5a1a1f6c7eef | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-056r-9500000000-0540950c816c3054bc75 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a70-9100000000-ab88b0c77d915bdf94c8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-05bb-8900000000-6145a4d3d74ad91e7c08 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4r-9500000000-e810b360131a0cd0626c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9000000000-fd760c42934208e53eab | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-05o0-3900000000-e52d00e2e7427df264f9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-05p9-9300000000-1e180faafcb2fce14102 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0bt9-9000000000-254ae2874ffc06d5337a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4s-4900000000-4b81a8a41abed679d134 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-9200000000-b73e0df104053da76268 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052f-9000000000-4d8ce26149e6d7a8763f | View in MoNA |
|---|
| MS | Mass Spectrum (Electron Ionization) | splash10-056r-9000000000-16517a02beb9427ceaff | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|