| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 14:21:11 -0600 |
|---|
| Update Date | 2015-06-03 17:19:03 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
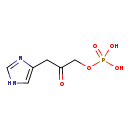
| Name: | Imidazole acetol-phosphate |
|---|
| Description | Imidazole acetol-phosphate is involved in the histidine biosynthesis pathway. Imidazole acetol-phosphate is created by the breakdown of D-erythro-imidazole-glycerol-phosphate into imidazole acetol-phosphate and H2O. Imidazoleglycerol-phosphate dehydratase catalyzes this reaction. |
|---|
| Structure | |
|---|
| Synonyms: | - 1-(1H-Imidazol-4-yl)-3-(phosphonooxy)-2-propanone
- 3-(1H-Imidazol-4-yl)-2-oxopropyl dihydrogen phosphate
- 3-(1H-Imidazol-4-yl)-2-oxopropyl dihydrogen phosphoric acid
- 3-(Imidazol-4-yl)-2-oxopropyl dihydrogen phosphate
- 3-(Imidazol-4-yl)-2-oxopropyl dihydrogen phosphoric acid
- 3-(Imidazol-4-yl)-2-oxopropyl phosphate
- 3-(Imidazol-4-yl)-2-oxopropyl phosphoric acid
- IAP
- Imidazole acetol phosphate
- Imidazole acetol phosphoric acid
- Imidazole acetol-p
- Imidazole acetol-phosphoric acid
- Imidazole-acetol phosphate
- Imidazole-acetol phosphoric acid
|
|---|
| Chemical Formula: | C6H9N2O5P |
|---|
| Weight: | Average: 220.1198
Monoisotopic: 220.02490792 |
|---|
| InChI Key: | YCFFMSOLUMRAMD-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/C6H9N2O5P/c9-6(3-13-14(10,11)12)1-5-2-7-4-8-5/h2,4H,1,3H2,(H,7,8)(H2,10,11,12) |
|---|
| CAS number: | 99979-59-6 |
|---|
| IUPAC Name: | [3-(1H-imidazol-4-yl)-2-oxopropoxy]phosphonic acid |
|---|
| Traditional IUPAC Name: | 3-(1H-imidazol-4-yl)-2-oxopropoxyphosphonic acid |
|---|
| SMILES: | OP(O)(=O)OCC(=O)CC1=CNC=N1 |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as monoalkyl phosphates. These are organic compounds containing a phosphate group that is linked to exactly one alkyl chain. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Organic phosphoric acids and derivatives |
|---|
| Sub Class | Phosphate esters |
|---|
| Direct Parent | Monoalkyl phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Monoalkyl phosphate
- Azole
- Imidazole
- Heteroaromatic compound
- Ketone
- Organoheterocyclic compound
- Azacycle
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Organopnictogen compound
- Organic oxygen compound
- Carbonyl group
- Organic nitrogen compound
- Hydrocarbon derivative
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -2 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | |
|---|
| KEGG Pathways: | - Histidine metabolism ec00340
- Metabolic pathways eco01100
- Novobiocin biosynthesis ec00401
- Phenylalanine metabolism ec00360
- Phenylalanine, tyrosine and tryptophan biosynthesis ec00400
- Tropane, piperidine and pyridine alkaloid biosynthesis ec00960
- Tyrosine metabolism ec00350
|
|---|
| EcoCyc Pathways: | |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-000t-9200000000-41a681dee161ad4f3e7b | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0fk9-3390000000-102fb9b517f2001e0996 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0kmi-4950000000-076be23324f875b841dc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a59-9500000000-99bd6a8eac665bc00868 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-016r-7190000000-c52bfb05c2cd90023536 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9000000000-7598c15214a6b2b67038 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-59a5e26dad504ee2412d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-0090000000-199895f882e036c55087 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-3930000000-3a2cf619e974119e8f24 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0pw9-9300000000-f31212a9e65f18dabcb3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-2090000000-bd2937021ad7bda9fa53 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9020000000-af7370fa1a56f74a6fb4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-8c5fbfcdabecdcd6d55b | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Albritton WL, Levin AP: Some comparative kinetic data on the enzyme imidazoleacetol phosphate:L-glutamate aminotransferase derived from mutant strains of Salmonella typhimurium. J Biol Chem. 1970 May 25;245(10):2525-8. Pubmed: 5445798
- AMES BN, HORECKER BL: The biosynthesis of histidine: imidazoleacetol phosphate transaminase. J Biol Chem. 1956 May;220(1):113-28. Pubmed: 13319331
- AMES BN, MITCHELL HK: The biosynthesis of histidine; imidazoleglycerol phosphate, imidazoleacetol phosphate, and histidinol phosphate. J Biol Chem. 1955 Feb;212(2):687-96. Pubmed: 14353870
- Henderson GB, Snell EE: Vitamin B 6 -responsive histidine deficiency in mutants of Salmonella typhimurium. Proc Natl Acad Sci U S A. 1971 Dec;68(12):2903-7. Pubmed: 4943547
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- LEVIN AP, HARTMAN PE: ACTION OF A HISTIDINE ANALOGUE, 1,2,4-TRIAZOLE-3-ALANINE, IN SALMONELLA TYPHIMURIUM. J Bacteriol. 1963 Oct;86:820-8. Pubmed: 14066480
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|