Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
Adenosyl cobinamide phosphate (M2MDB000825)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-05-31 14:20:52 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 17:19:02 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Adenosyl cobinamide phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Adenosyl cobinamide phosphate, a known intermediate of the de novo biosynthetic pathway, is involved in Porphyrin and chlorophyll metabolism.In Salmonella typhimurium LT2, under anaerobic conditions, CobU (EC 2.7.7.62 and EC 2.7.1.156), CobT (EC 2.4.2.21), CobC (EC 3.1.3.73) and CobS (EC 2.7.8.26) catalyse reactions in the nucleotide loop | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
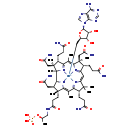
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C58H85CoN16O14P | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 1320.3013 Monoisotopic: 1319.550078214 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | MQCMBMUJJHSGIF-QMUWONGRSA-M | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C48H74N11O11P.C10H12N5O3.Co/c1-23(70-71(67,68)69)22-55-38(66)16-17-45(6)29(18-35(52)63)43-48(9)47(8,21-37(54)65)28(12-15-34(51)62)40(59-48)25(3)42-46(7,20-36(53)64)26(10-13-32(49)60)30(56-42)19-31-44(4,5)27(11-14-33(50)61)39(57-31)24(2)41(45)58-43;1-4-6(16)7(17)10(18-4)15-3-14-5-8(11)12-2-13-9(5)15;/h19,23,26-29,43H,10-18,20-22H2,1-9H3,(H16,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69);2-4,6-7,10,16-17H,1H2,(H2,11,12,13);/q;;+2/p-1/t23-,26-,27-,28-,29+,43-,45-,46+,47+,48+;4-,6-,7-,10-;/m11./s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (3R,4S,5S,9S,10S,15S,19R,20R,21R)-1-{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl}-5,10,15-tris(2-carbamoylethyl)-4,9,20-tris(carbamoylmethyl)-3,4,7,9,14,14,17,19-octamethyl-19-(2-{[(2R)-2-(phosphonooxy)propyl]carbamoyl}ethyl)-2lambda5,22,23lambda5,24lambda5-tetraaza-1-cobaltaoctacyclo[11.9.1.1^{1,8}.0^{2,6}.0^{3,21}.0^{16,23}.0^{18,22}.0^{11,24}]tetracosa-2(6),7,11(24),12,16(23),17-hexaene-2,23,24-tris(ylium)-1,1-diuide | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | (3R,4S,5S,9S,10S,15S,19R,20R,21R)-1-{[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl}-5,10,15-tris(2-carbamoylethyl)-4,9,20-tris(carbamoylmethyl)-3,4,7,9,14,14,17,19-octamethyl-19-(2-{[(2R)-2-(phosphonooxy)propyl]carbamoyl}ethyl)-2lambda5,22,23lambda5,24lambda5-tetraaza-1-cobaltaoctacyclo[11.9.1.1^{1,8}.0^{2,6}.0^{3,21}.0^{16,23}.0^{18,22}.0^{11,24}]tetracosa-2(6),7,11(24),12,16(23),17-hexaene-2,23,24-tris(ylium)-1,1-diuide | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | C[C@H](CNC(=O)CC[C@]1(C)[C@@H](CC(=O)N)[C@@]2([H])N([Co+]C[C@H]3O[C@H]([C@H](O)[C@@H]3O)N3C=NC4=C3N=CN=C4N)\C1=C(C)/C1=N/C(=C\C3=N\C(=C(C)/C4=N[C@]2(C)[C@@](C)(CC(=O)N)[C@@H]4CCC(=O)N)\[C@@](C)(CC(=O)N)[C@@H]3CCC(=O)N)/C(C)(C)[C@@H]1CCC(=O)N)OP(=O)(O)O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as metallotetrapyrroles. These are polycyclic compounds containing a tetrapyrrole skeleton combined with a metal atom. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organoheterocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Tetrapyrroles and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Metallotetrapyrroles | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Metallotetrapyrroles | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosyl cobinamide + Adenosine triphosphate > Adenosyl cobinamide phosphate + ADP + Hydrogen ion Adenosyl cobinamide phosphate + Guanosine triphosphate + Hydrogen ion > Adenosylcobinamide-GDP + Pyrophosphate Adenosyl cobinamide + Adenosine triphosphate <> Adenosyl cobinamide phosphate + ADP Adenosyl cobinamide phosphate + Guanosine triphosphate <> Adenosylcobinamide-GDP + Pyrophosphate Adenosyl cobinamide + Guanosine triphosphate <> Adenosyl cobinamide phosphate + Guanosine diphosphate Adenosine triphosphate + Guanosine triphosphate + Adenosyl cobinamide <> Adenosyl cobinamide phosphate + ADP + Guanosine diphosphate Adenosyl cobinamide + Adenosine triphosphate > Adenosyl cobinamide phosphate + ADP + Hydrogen ion Adenosyl cobinamide phosphate + Guanosine triphosphate + Hydrogen ion > Adenosylcobinamide-GDP + Pyrophosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in nucleotide binding
- Specific function:
- ATP-dependent phosphorylation of adenosylcobinamide and adds GMP to adenosylcobinamide phosphate
- Gene Name:
- cobU
- Uniprot ID:
- P0AE76
- Molecular weight:
- 20164
Reactions
| ATP or GTP + adenosylcobinamide = adenosylcobinamide phosphate + ADP or GDP. |
| GTP + adenosylcobinamide phosphate = diphosphate + adenosylcobinamide-GDP. |