Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
2-Isopropyl-3-oxosuccinate (M2MDB000823)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-05-31 14:20:46 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 17:19:02 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | 2-Isopropyl-3-oxosuccinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | 2-Isopropyl-3-oxosuccinate is an intermediate in leucine biosynthesis and can be generated from (2R,3S)-3-Isopropylmalate. It is the third step in leucine biosynthesis after the fork from valine synthesis. It is an oxidative decarboxylation. Leucine biosynthesis involves a five-step conversion process starting with the valine precursor 2-keto-isovalerate. The final step in this pathway is catalyzed by two transaminases of broad specificity, Branched-chain amino acid transferase (IlvE) and Tyrosine aminotransferase (TyrB). In this pathway 2-Isopropyl-3-oxosuccinate is converted to 4-Methyl-2-oxopentanoate via spontaneous reaction.(BioCyc) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
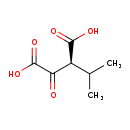
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C7H10O5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 174.1513 Monoisotopic: 174.05282343 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | HIIZAGQWABAMRR-BYPYZUCNSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C7H10O5/c1-3(2)4(6(9)10)5(8)7(11)12/h3-4H,1-2H3,(H,9,10)(H,11,12)/t4-/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (3S)-2-oxo-3-(propan-2-yl)butanedioic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | (2S)-2-isopropyl-3-oxobutanedioic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | CC(C)[C@H](C(O)=O)C(=O)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as short-chain keto acids and derivatives. These are keto acids with an alkyl chain the contains less than 6 carbon atoms. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Keto acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Short-chain keto acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Short-chain keto acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | 3-Isopropylmalate + NAD > 2-Isopropyl-3-oxosuccinate + Hydrogen ion + NADH 2-Isopropyl-3-oxosuccinate + Hydrogen ion > Ketoleucine + Carbon dioxide 3-Isopropylmalate + NAD <> 2-Isopropyl-3-oxosuccinate + NADH + Hydrogen ion 3-Isopropylmalate + NAD + 2-Isopropyl-3-oxosuccinate <> Ketoleucine + Carbon dioxide + NADH + Hydrogen ion 3 3-Isopropylmalate + NAD >2 2-Isopropyl-3-oxosuccinate + Hydrogen ion + NADH 3 3-Isopropylmalate + NAD >2 2-Isopropyl-3-oxosuccinate + Hydrogen ion + NADH | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in magnesium ion binding
- Specific function:
- Catalyzes the oxidation of 3-carboxy-2-hydroxy-4- methylpentanoate (3-isopropylmalate) to 3-carboxy-4-methyl-2- oxopentanoate. The product decarboxylates to 4-methyl-2 oxopentanoate
- Gene Name:
- leuB
- Uniprot ID:
- P30125
- Molecular weight:
- 39517
Reactions
| (2R,3S)-3-isopropylmalate + NAD(+) = 4-methyl-2-oxopentanoate + CO(2) + NADH. |
- General function:
- Involved in magnesium ion binding
- Specific function:
- Catalyzes the NAD(+)-dependent oxidative decarboxylation of D-malate into pyruvate. Is essential for aerobic growth on D- malate as the sole carbon source. But is not required for anaerobic D-malate utilization, although DmlA is expressed and active in those conditions. Appears to be not able to use L- tartrate as a substrate for dehydrogenation instead of D-malate
- Gene Name:
- dmlA
- Uniprot ID:
- P76251
- Molecular weight:
- 40315
Reactions
| (R)-malate + NAD(+) = pyruvate + CO(2) + NADH. |