Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
Tartronate semialdehyde (M2MDB000707)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-05-31 14:10:18 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 15:54:57 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Tartronate semialdehyde | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Tartronate semialdehyde is an intermediate in ascorbate and aldarate as well as glyoxylate and dicarboxylate metabolism. It is generated from 2-dehydro-3-deoxy-D-glucarate and 5-dehydro-4-deoxy-D-glucarate via the enzyme 2-dehydro-3-deoxyglucarate aldolase [EC:4.1.2.20]. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C3H4O4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 104.0615 Monoisotopic: 104.010958616 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | QWBAFPFNGRFSFB-UHFFFAOYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C3H4O4/c4-1-2(5)3(6)7/h1-2,5H,(H,6,7) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | 2-hydroxy-3-oxopropanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | tartronate semialdehyde | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
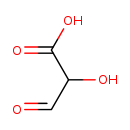
| SMILES: | OC(C=O)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as monosaccharides. Monosaccharides are compounds containing one carbohydrate unit not glycosidically linked to another such unit, and no set of two or more glycosidically linked carbohydrate units. Monosaccharides have the general formula CnH2nOn. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic oxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Carbohydrates and carbohydrate conjugates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Monosaccharides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Tartronate semialdehyde + Hydrogen ion + NADH <> Glyceric acid + NAD 2 Glyoxylic acid + Hydrogen ion <> Tartronate semialdehyde + Carbon dioxide Hydroxypyruvic acid <> Tartronate semialdehyde 5-Dehydro-4-deoxy-D-glucarate > Tartronate semialdehyde + Pyruvic acid 2 Glyoxylic acid <> Tartronate semialdehyde + Carbon dioxide Glyceric acid + NAD <> Tartronate semialdehyde + NADH + Hydrogen ion Glyceric acid + NADP <> Tartronate semialdehyde + NADPH + Hydrogen ion 5-Dehydro-4-deoxy-D-glucarate <> Pyruvic acid + Tartronate semialdehyde Tartronate semialdehyde + Pyruvic acid <> 2-Dehydro-3-deoxy-D-glucarate Hydrogen ion + Glyoxylic acid > Carbon dioxide + Tartronate semialdehyde NAD(P)<sup>+</sup> + Glyceric acid < NAD(P)H + Tartronate semialdehyde + Hydrogen ion 2-Dehydro-3-deoxy-D-glucarate > Pyruvic acid + Tartronate semialdehyde Glyceric acid + NAD(P)(+) > Tartronate semialdehyde + NAD(P)H 2 Glyoxylic acid > Tartronate semialdehyde + Carbon dioxide Hydroxypyruvic acid > Tartronate semialdehyde Glyceric acid + NAD + NADP <> Tartronate semialdehyde + NADH + NADPH + Hydrogen ion 5-dehydro-4-deoxy-D-glucarate(2−) > Pyruvic acid + Tartronate semialdehyde Tartronate semialdehyde + Hydrogen ion + NADPH + NADPH > NADP + Glyceric acid 2 Glyoxylic acid + Hydrogen ion > Carbon dioxide + Tartronate semialdehyde Tartronate semialdehyde + NADH + Hydrogen ion > NAD + Glyceric acid Tartronate semialdehyde + Hydrogen ion + NADH <> Glyceric acid + NAD Hydroxypyruvic acid <> Tartronate semialdehyde 2 Glyoxylic acid + Hydrogen ion <> Tartronate semialdehyde + Carbon dioxide Hydroxypyruvic acid <> Tartronate semialdehyde | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in 3-hydroxyisobutyrate dehydrogenase activity
- Specific function:
- (R)-glycerate + NAD(P)(+) = 2-hydroxy-3- oxopropanoate + NAD(P)H
- Gene Name:
- garR
- Uniprot ID:
- P0ABQ2
- Molecular weight:
- 30427
Reactions
| D-glycerate + NAD(P)(+) = 2-hydroxy-3-oxopropanoate + NAD(P)H. |
- General function:
- Involved in tartronate-semialdehyde synthase activity
- Specific function:
- Catalyzes the condensation of two molecules of glyoxylate to give 2-hydroxy-3-oxopropanoate (also termed tartronate semialdehyde)
- Gene Name:
- gcl
- Uniprot ID:
- P0AEP7
- Molecular weight:
- 64731
Reactions
| 2 glyoxylate = tartronate semialdehyde + CO(2). |
- General function:
- Involved in carbon-carbon lyase activity
- Specific function:
- Catalyzes the reversible retro-aldol cleavage of both 5- keto-4-deoxy-D-glucarate and 2-keto-3-deoxy-D-glucarate to pyruvate and tartronic semialdehyde
- Gene Name:
- garL
- Uniprot ID:
- P23522
- Molecular weight:
- 27384
Reactions
| 5-dehydro-4-deoxy-D-glucarate = pyruvate + tartronate semialdehyde. |
| 2-dehydro-3-deoxy-D-glucarate = pyruvate + tartronate semialdehyde. |
- General function:
- Carbohydrate transport and metabolism
- Specific function:
- Catalyzes the reversible isomerization between hydroxypyruvate and 2-hydroxy-3-oxopropanoate (also termed tartronate semialdehyde)
- Gene Name:
- hyi
- Uniprot ID:
- P30147
- Molecular weight:
- 29377
Reactions
| Hydroxypyruvate = 2-hydroxy-3-oxopropanoate. |
- General function:
- Involved in 3-hydroxyisobutyrate dehydrogenase activity
- Specific function:
- (R)-glycerate + NAD(P)(+) = 2-hydroxy-3- oxopropanoate + NAD(P)H
- Gene Name:
- glxR
- Uniprot ID:
- P77161
- Molecular weight:
- 30800
Reactions
| D-glycerate + NAD(P)(+) = 2-hydroxy-3-oxopropanoate + NAD(P)H. |