Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
3-Dehydro-L-gulonate (M2MDB000665)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-05-31 14:08:05 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 15:54:51 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | 3-Dehydro-L-gulonate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | 3-Dehydro-L-gulonate is an intermediate in ascorbic acid degradation. It is a substrate for the enzyme 2,3-diketo-L-gulonate reductase. This enzyme catalyzes the reduction of 2,3-diketo-L-gulonate in the presence of NADH, to form 3-keto-L-gulonate. It participates in the reaction: 3-dehydro-L-gulonate + NAD(P)+ = (4R,5S)-4,5,6-trihydroxy-2,3-dioxohexanoate + NAD(P)H. Several pathways for the irreversible catabolism of ascorbate have been described. Facultatively aerobic bacteria such as Escherichia coli degrade L-ascorbate by different pathways under aerobic and anaerobic conditions. The anaerobic pathway begins with phosphorylation of ascorbate (mediated by a PTS-type transporter), while the aerobic pathway proceeds via 2,3-dioxo-L-gulonate. Both pathways produce D-xylulose 5-phosphate, a centeral metabolite that is fed into the pentose phosphate pathway. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
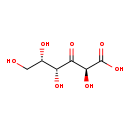
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C6H10O7 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 194.1394 Monoisotopic: 194.042652674 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | WTAHRPBPWHCMHW-LWKDLAHASA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C6H10O7/c7-1-2(8)3(9)4(10)5(11)6(12)13/h2-3,5,7-9,11H,1H2,(H,12,13)/t2-,3+,5-/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2S,4R,5S)-2,4,5,6-tetrahydroxy-3-oxohexanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | 3-dehydro-L-gulonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | OC[C@H](O)[C@@H](O)C(=O)[C@H](O)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as sugar acids and derivatives. Sugar acids and derivatives are compounds containing a saccharide unit which bears a carboxylic acid group. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic oxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Carbohydrates and carbohydrate conjugates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Sugar acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | 2,3-Diketo-L-gulonate + Hydrogen ion + NADH > 3-Dehydro-L-gulonate + NAD 3-Dehydro-L-gulonate + Adenosine triphosphate > 3-Dehydro-L-gulonate 6-phosphate + ADP + Hydrogen ion 3-Dehydro-L-gulonate + NAD <> 2,3-Diketo-L-gulonate + NADH + Hydrogen ion 3-Dehydro-L-gulonate + NADP <> 2,3-Diketo-L-gulonate + NADPH + Hydrogen ion 3-Dehydro-L-gulonate + Adenosine triphosphate <> 3-Dehydro-L-gulonate 6-phosphate + ADP 3-Dehydro-L-gulonate + NAD < Hydrogen ion + 2,3-Diketo-L-gulonate + NADH 3-Dehydro-L-gulonate + NAD(P)(+) > (4R,5S)-4,5,6-trihydroxy-2,3-dioxohexanoate + NAD(P)H Adenosine triphosphate + 3-Dehydro-L-gulonate > ADP + 3-Dehydro-L-gulonate 6-phosphate 3-Dehydro-L-gulonate + NAD + NADP <> 2,3-Diketo-L-gulonate + NADH + NADPH + Hydrogen ion 3-Dehydro-L-gulonate + Adenosine triphosphate > 3-keto-L-gulonate 6-phosphate + Adenosine diphosphate + Hydrogen ion + 3-Keto-L-gulonate 6-phosphate + ADP 2,3-Diketo-L-gulonate + NADH + Hydrogen ion + 2,3-Diketo-L-gulonate > 3-Dehydro-L-gulonate + NAD | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor
- Specific function:
- Catalyzes the reduction of 2,3-diketo-L-gulonate in the presence of NADH, to form 3-keto-L-gulonate
- Gene Name:
- dlgD
- Uniprot ID:
- P37672
- Molecular weight:
- 36572
Reactions
| 3-dehydro-L-gulonate + NAD(P)(+) = (4R,5S)-4,5,6-trihydroxy-2,3-dioxohexanoate + NAD(P)H. |
- General function:
- Involved in phosphotransferase activity, alcohol group as acceptor
- Specific function:
- Catalyzes the phosphorylation of L-xylulose and 3-keto- L-gulonate. Is involved in L-lyxose utilization via xylulose, and may also be involved in the utilization of 2,3-diketo-L-gulonate
- Gene Name:
- lyx
- Uniprot ID:
- P37677
- Molecular weight:
- 55155
Reactions
| ATP + L-xylulose = ADP + L-xylulose 5-phosphate. |
| ATP + 3-dehydro-L-gulonate = ADP + 3-dehydro-L-gulonate 6-phosphate. |