| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 14:03:51 -0600 |
|---|
| Update Date | 2015-06-03 15:54:41 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
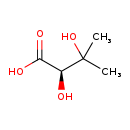
| Name: | (R)-2,3-Dihydroxy-isovalerate |
|---|
| Description | (r)-2,3-dihydroxy-isovalerate belongs to the class of Branched Fatty Acids. These are fatty acids containing a branched chain. (inferred from compound structure) |
|---|
| Structure | |
|---|
| Synonyms: | - α,β-dihydroxy-isovalerate
- α,β-dihydroxy-isovaleric acid
- (2R)-2,3-dihydroxy-3-methylbutanoate
- (2R)-2,3-dihydroxy-3-methylbutanoic acid
- (R)-2,3-dihydroxy-3-methylbutanoate
- (R)-2,3-dihydroxy-3-methylbutanoic acid
- (R)-2,3-Dihydroxy-isovaleric acid
- 2,3-dihydroxy-isovalerate
- 2,3-Dihydroxy-isovaleric acid
|
|---|
| Chemical Formula: | C5H10O4 |
|---|
| Weight: | Average: 134.1305
Monoisotopic: 134.057908808 |
|---|
| InChI Key: | JTEYKUFKXGDTEU-VKHMYHEASA-N |
|---|
| InChI: | InChI=1S/C5H10O4/c1-5(2,9)3(6)4(7)8/h3,6,9H,1-2H3,(H,7,8)/t3-/m0/s1 |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | (2R)-2,3-dihydroxy-3-methylbutanoic acid |
|---|
| Traditional IUPAC Name: | (R)-2,3-dihydroxy-isovalerate |
|---|
| SMILES: | CC(C)(O)[C@@H](O)C(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as hydroxy fatty acids. These are fatty acids in which the chain bears a hydroxyl group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acids and conjugates |
|---|
| Direct Parent | Hydroxy fatty acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Branched fatty acid
- Methyl-branched fatty acid
- Short-chain hydroxy acid
- Hydroxy fatty acid
- Alpha-hydroxy acid
- Hydroxy acid
- Monosaccharide
- Tertiary alcohol
- 1,2-diol
- Secondary alcohol
- Carboxylic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic oxygen compound
- Carbonyl group
- Hydrocarbon derivative
- Organic oxide
- Alcohol
- Organooxygen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -1 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | (R)-2,3-Dihydroxy-isovalerate > alpha-Ketoisovaleric acid + Water(R)-2,3-Dihydroxy-isovalerate + NADP <> (S)-2-Acetolactate + Hydrogen ion + NADPH(R)-2,3-Dihydroxy-isovalerate + NADP <> 3-Hydroxy-3-methyl-2-oxobutanoic acid + NADPH + Hydrogen ion(R)-2,3-Dihydroxy-isovalerate <> alpha-Ketoisovaleric acid + Water(R)-2,3-Dihydroxy-isovalerate + NADP <> (<i>S</i>)-2-acetolactate + NADPH + Hydrogen ion(R)-2,3-Dihydroxy-isovalerate + NADP > (S)-2-Acetolactate + NADPH(R)-2,3-Dihydroxy-isovalerate + Hydrogen ion + NADPH + NADPH > NADP + (R) 2,3-Dihydroxy-3-methylvalerate(S)-2-Acetolactate + Hydrogen ion + NADPH + NADPH > NADP + (R)-2,3-Dihydroxy-isovalerate(R)-2,3-Dihydroxy-isovalerate > Water + Isovaleric acid(R)-2,3-Dihydroxy-isovalerate > alpha-Ketoisovaleric acid + Water |
|---|
| SMPDB Pathways: | |
|---|
| KEGG Pathways: | - Metabolic pathways eco01100
- Pantothenate and CoA biosynthesis ec00770
- Valine, leucine and isoleucine biosynthesis ec00290
|
|---|
| EcoCyc Pathways: | |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0a4i-9000000000-59e6990530d950227520 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-00s9-9473000000-9708bfc20ee5ae043678 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00kr-4900000000-196ffbbf313a33f24f27 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00ri-9300000000-aebebbbe2c7c11465a67 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-92af6a04d8cb7601ab35 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0019-9500000000-2837622949a6bd688094 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00s9-9400000000-0fc7aa81197fc5feb444 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00dr-9000000000-45f3b95b2f30ea8445c1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-2900000000-fab4ee9aeca7067a9207 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-056r-9100000000-b3da44872fd12f996ec6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05fr-9000000000-571b504e001a1456ab10 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00kr-9600000000-e4a123bb359dea0126e3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-05g0-9000000000-30fdda4eee690af3ee50 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-285f7931bf8163b701da | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- Yurtsever D. (2007). Fatty acid methyl ester profiling of Enterococcus and Esherichia coli for microbial source tracking. M.sc. Thesis. Villanova University: U.S.A
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|