Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
2-Dehydro-D-gluconate (M2MDB000575)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-05-31 14:03:06 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-09-17 15:41:46 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | 2-Dehydro-D-gluconate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | 2-Keto-L-gluconate is a derivative of gluconic acid, which occurs naturally in fruit, honey and wine and is used as a food additive, an acidity regulator. It is also used in cleaning products where it helps cleaning up mineral deposits. It is a strong chelating agent, especially in alkaline solution. It chelates the anions of calcium, iron, aluminium, copper, and other heavy metals. E. coli contains ketogluconate reductase genes, so it is likely that pathways exist for the metabolism of ketogluconate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C6H10O7 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 194.1394 Monoisotopic: 194.042652674 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | VBUYCZFBVCCYFD-UHFFFAOYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C6H10O7/c7-1-2(8)3(9)4(10)5(11)6(12)13/h2-4,7-10H,1H2,(H,12,13) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (3S,4R,5R)-3,4,5,6-tetrahydroxy-2-oxohexanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | 2-ketogluconic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
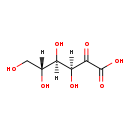
| SMILES: | OCC(O)C(O)C(O)C(=O)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as sugar acids and derivatives. Sugar acids and derivatives are compounds containing a saccharide unit which bears a carboxylic acid group. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic oxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Carbohydrates and carbohydrate conjugates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Sugar acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 157.82 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Gluconic acid + NADP <> 2-Keto-D-gluconic acid + NADPH + Hydrogen ion + 2-Dehydro-D-gluconate NADP + Gluconic acid <> Hydrogen ion + 2-Dehydro-D-gluconate + NADPH Hydrogen ion + NADPH + 2,5-Diketo-D-gluconate <> 2-Dehydro-D-gluconate + NADP Hydrogen ion + 2-Dehydro-D-gluconate + NADPH <> L-Idonate + NADP 2-Dehydro-D-gluconate + NADP > 2,5-didehydro-D-gluconate + NADPH Gluconic acid + NADP > 2-Dehydro-D-gluconate + NADPH 2-Dehydro-D-gluconate + NADP <> 2,5-Diketo-D-gluconate + NADPH + Hydrogen ion 2-Keto-L-gluconate + Hydrogen ion + NADPH + 2-Dehydro-D-gluconate + NADPH > NADP + L-Idonate 2,5-Diketo-D-gluconate + Hydrogen ion + NADPH + NADPH > NADP + 2-Keto-L-gluconate + 2-Dehydro-D-gluconate 2-Keto-L-gluconate + NADPH + Hydrogen ion + 2-Dehydro-D-gluconate + NADPH > Gluconic acid + NADP 2 2-Dehydro-D-gluconate + NADP <>2 2,5-Diketo-D-gluconate + NADPH + Hydrogen ion | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in oxidoreductase activity
- Specific function:
- Catalyzes the reduction of 2,5-diketo-D-gluconic acid (25DKG) to 2-keto-L-gulonic acid (2KLG)
- Gene Name:
- dkgB
- Uniprot ID:
- P30863
- Molecular weight:
- 29437
Reactions
| 2-dehydro-D-gluconate + NADP(+) = 2,5-didehydro-D-gluconate + NADPH. |
- General function:
- Involved in oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor
- Specific function:
- Catalyzes the NADPH-dependent reduction of glyoxylate and hydroxypyruvate into glycolate and glycerate, respectively. Can also reduce 2,5-diketo-D-gluconate (25DKG) to 5-keto-D- gluconate (5KDG), 2-keto-D-gluconate (2KDG) to D-gluconate, and 2- keto-L-gulonate (2KLG) to L-idonate (IA), but it is not its physiological function. Inactive towards 2-oxoglutarate, oxaloacetate, pyruvate, 5-keto-D-gluconate, D-fructose and L- sorbose. Activity with NAD is very low
- Gene Name:
- ghrB
- Uniprot ID:
- P37666
- Molecular weight:
- 35395
Reactions
| Glycolate + NADP(+) = glyoxylate + NADPH. |
| D-glycerate + NAD(P)(+) = hydroxypyruvate + NAD(P)H. |
| D-gluconate + NADP(+) = 2-dehydro-D-gluconate + NADPH. |
- General function:
- Involved in oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor
- Specific function:
- Catalyzes the reduction of 2,3-diketo-L-gulonate in the presence of NADH, to form 3-keto-L-gulonate
- Gene Name:
- dlgD
- Uniprot ID:
- P37672
- Molecular weight:
- 36572
Reactions
| 3-dehydro-L-gulonate + NAD(P)(+) = (4R,5S)-4,5,6-trihydroxy-2,3-dioxohexanoate + NAD(P)H. |
- General function:
- Involved in oxidoreductase activity
- Specific function:
- Catalyzes the reduction of 2,5-diketo-D-gluconic acid (25DKG) to 2-keto-L-gulonic acid (2KLG). It is also capable of stereoselective -keto ester reductions on ethyl acetoacetate and other 2-substituted derivatives
- Gene Name:
- dkgA
- Uniprot ID:
- Q46857
- Molecular weight:
- 31109
Reactions
| 2-dehydro-D-gluconate + NADP(+) = 2,5-didehydro-D-gluconate + NADPH. |