| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 13:59:06 -0600 |
|---|
| Update Date | 2015-06-03 15:54:23 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | Pantetheine |
|---|
| Description | Pantetheine is the mercaptoethyl conjugated amide analogue of pantothenic acid (Vitamin B5). The dimer of this compound, pantethine is more commonly known, and is considered to be a more potent form of vitamin B5 than pantothenic acid. Pantetheine is an intermediate in the production of coenzyme A, an intermediate in the pathway of coenzyme A formation. |
|---|
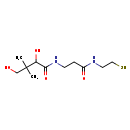
| Structure | |
|---|
| Synonyms: | - (R)-Pantetheine
- D-Pantetheine
- Lactobacillus bulgaricus factor
- LBF
- Pantotheine
|
|---|
| Chemical Formula: | C11H22N2O4S |
|---|
| Weight: | Average: 278.368
Monoisotopic: 278.130027892 |
|---|
| InChI Key: | ZNXZGRMVNNHPCA-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/C11H22N2O4S/c1-11(2,7-14)9(16)10(17)13-4-3-8(15)12-5-6-18/h9,14,16,18H,3-7H2,1-2H3,(H,12,15)(H,13,17) |
|---|
| CAS number: | 496-65-1 |
|---|
| IUPAC Name: | 2,4-dihydroxy-3,3-dimethyl-N-{2-[(2-sulfanylethyl)carbamoyl]ethyl}butanamide |
|---|
| Traditional IUPAC Name: | pantetheine |
|---|
| SMILES: | CC(C)(CO)C(O)C(=O)NCCC(=O)NCCS |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as beta amino acids and derivatives. These are amino acids having a (-NH2) group attached to the beta carbon atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Beta amino acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Beta amino acid or derivatives
- Fatty amide
- Monosaccharide
- N-acyl-amine
- Fatty acyl
- Carboxamide group
- Secondary alcohol
- Secondary carboxylic acid amide
- Alkylthiol
- Primary alcohol
- Organosulfur compound
- Organooxygen compound
- Organonitrogen compound
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Alcohol
- Carbonyl group
- Organic nitrogen compound
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | 0 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | | Pantothenate and CoA biosynthesis | PW000828 | 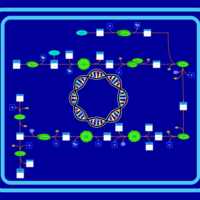 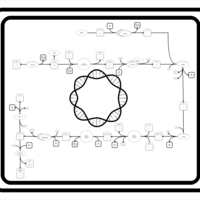 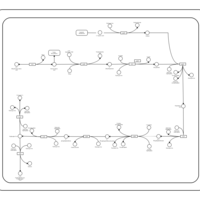 |
|
|---|
| KEGG Pathways: | - Pantothenate and CoA biosynthesis ec00770
|
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0irr-9850000000-e4fcca3c68a52e72a3da | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-00di-9522200000-170f850fbccf97bf4dfb | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01t9-5690000000-a5cb9d4433d26f7a2ac0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01t9-9410000000-895050a718b98df8818e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0hbc-9200000000-c3a563dd190af4f15252 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-2390000000-5ca6485cda20e56cabaf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03ka-6950000000-b90e9c6f3732fa15365a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00fr-9300000000-12baf4bf4d07fb8992b4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0190000000-74102e0d21631b5f176c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-2900000000-8b6fdd5f7342396d976f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00kg-8910000000-5a7f4294d5361d3252db | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0290000000-e6596ba35ea7b2d6a1bb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-004j-6930000000-cd8c5ecad3eccc49fbb7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-004i-9400000000-9b89efff76cf7f21adb4 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
|
|---|
| Synthesis Reference: | Mandel, Alexander L.; La Clair, James J.; Burkart, Michael D. Modular Synthesis of Pantetheine and Phosphopantetheine. Organic Letters (2004), 6(26), 4801-4803. |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|