| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 13:53:13 -0600 |
|---|
| Update Date | 2015-09-13 12:56:11 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
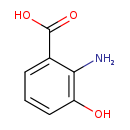
| Name: | 3-Hydroxyanthranilic acid |
|---|
| Description | 3-Hydroxyanthranilic acid is an oxidation product of tryptophan metabolism. It may be a free radical scavenger and a carcinogen. |
|---|
| Structure | |
|---|
| Synonyms: | - 2-Amino-3-hydroxy-Benzoate
- 2-Amino-3-hydroxy-Benzoic acid
- 2-Amino-3-hydroxybenzoate
- 2-Amino-3-hydroxybenzoic acid
- 3-Hydroxanthranilate
- 3-Hydroxanthranilic acid
- 3-Hydroxy-2-aminobenzoate
- 3-Hydroxy-2-aminobenzoic acid
- 3-Hydroxy-Anthranilate
- 3-Hydroxy-Anthranilic acid
- 3-Hydroxy-anthranilsaeure
- 3-Hydroxyanthranilate
- 3-Hydroxyanthranilic acid
- 3-Hydroxyantranilate
- 3-Hydroxyantranilic acid
- 3-OH-anthranilate
- 3-OH-anthranilic acid
- 3-OHAA
- 3-Oxyanthranilate
- 3-Oxyanthranilic acid
|
|---|
| Chemical Formula: | C7H7NO3 |
|---|
| Weight: | Average: 153.1354
Monoisotopic: 153.042593095 |
|---|
| InChI Key: | WJXSWCUQABXPFS-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/C7H7NO3/c8-6-4(7(10)11)2-1-3-5(6)9/h1-3,9H,8H2,(H,10,11) |
|---|
| CAS number: | 548-93-6 |
|---|
| IUPAC Name: | 2-amino-3-hydroxybenzoic acid |
|---|
| Traditional IUPAC Name: | 3-hydroxyanthranilic acid |
|---|
| SMILES: | NC1=C(O)C=CC=C1C(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as hydroxybenzoic acid derivatives. Hydroxybenzoic acid derivatives are compounds containing a hydroxybenzoic acid (or a derivative), which is a benzene ring bearing a carboxyl and a hydroxyl groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Benzenoids |
|---|
| Class | Benzene and substituted derivatives |
|---|
| Sub Class | Benzoic acids and derivatives |
|---|
| Direct Parent | Hydroxybenzoic acid derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Aminobenzoic acid
- Aminobenzoic acid or derivatives
- Hydroxybenzoic acid
- Benzoic acid
- O-aminophenol
- Aminophenol
- Benzoyl
- Aniline or substituted anilines
- 1-hydroxy-4-unsubstituted benzenoid
- 1-hydroxy-2-unsubstituted benzenoid
- Phenol
- Vinylogous amide
- Amino acid or derivatives
- Amino acid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organonitrogen compound
- Hydrocarbon derivative
- Amine
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Organic nitrogen compound
- Organooxygen compound
- Primary amine
- Aromatic homomonocyclic compound
|
|---|
| Molecular Framework | Aromatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -1 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | |
|---|
| KEGG Pathways: | - 1,4-Dichlorobenzene degradation ec00627
- Microbial metabolism in diverse environments ec01120
- Tryptophan metabolism ec00380
|
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (Non-derivatized) | splash10-0zfv-0945000000-e3389ab25b7133eb184d | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (3 TMS) | splash10-0zfu-1955000000-fd51cc45bf09750e2b48 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0zfv-0945000000-e3389ab25b7133eb184d | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-0zfu-1955000000-fd51cc45bf09750e2b48 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0zfv-0934000000-11e2b38a36cb364bcaf3 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0006-0920000000-a57d310e9ca51aefd0c1 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0zg0-2900000000-f4f0b5c84821e81f2eea | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-00e9-6390000000-71ab4928435b01bf5b59 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-000i-0900000000-2313e347dd8659099393 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-001i-9400000000-4e342fdebc8145b606a1 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-001i-9000000000-ee5255b437ae762be1f9 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-qTof , Positive | splash10-000t-2900000000-a68dc7ca20e1f9595f15 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-0zfr-0900000000-ced0a738a11057bc94e4 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-0a4i-0900000000-cded3154e50c6169c670 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-0a4i-0900000000-9c388fc3035f19e3f156 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-0a4i-1900000000-9dbb06a7728aab173e0f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - , negative | splash10-0a4i-0900000000-19cfe2cda31cf7284669 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-000i-0900000000-02ae2bd70b54df77aca5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-000i-0900000000-56e21a4ececd5541a7d8 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-052r-0900000000-e3015612b3d263fabaa9 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-0a5i-3900000000-670c7d91a38e852d8e4c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - , positive | splash10-000i-5900000000-36b3a876d06807eb22a3 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 30V, Positive | splash10-052r-0900000000-e3015612b3d263fabaa9 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-0a5i-3900000000-670c7d91a38e852d8e4c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0900000000-56e21a4ececd5541a7d8 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0900000000-02ae2bd70b54df77aca5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-053i-3900000000-a410eeec27727c997dc7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0900000000-84ae4ad8e91e8e4b6a6b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0k9i-1900000000-6695f66ad1f45898b8b6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0pxr-9300000000-43045627b8c856bf07d2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0pb9-0900000000-810a4a12ecb151ecb516 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-1900000000-9f194909a88a4ac248b1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-8900000000-608d595690e7902b6195 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Calandra P: Research on tryptophan metabolites "via kynurenine" in epidermis of man and mouse. Acta Vitaminol Enzymol. 1975;29(1-6):158-60. Pubmed: 1244085
- De Antoni A, Rubaltelli FF, Costa C, Allegri G: Effect of phototherapy on the urinary excretion of tryptophan metabolites in neonatal hyperbilirubinemia. Acta Vitaminol Enzymol. 1975;29(1-6):145-50. Pubmed: 1244083
- Hegedus ZL, Frank HA, Altschule MD, Nayak U: Human plasma lipofuscin melanins formed from tryptophan metabolites. Arch Int Physiol Biochim. 1986 Dec;94(5):339-48. Pubmed: 2440410
- Herve C, Beyne P, Jamault H, Delacoux E: Determination of tryptophan and its kynurenine pathway metabolites in human serum by high-performance liquid chromatography with simultaneous ultraviolet and fluorimetric detection. J Chromatogr B Biomed Appl. 1996 Jan 12;675(1):157-61. Pubmed: 8634758
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Lopez AS, Alegre E, LeMaoult J, Carosella E, Gonzalez A: Regulatory role of tryptophan degradation pathway in HLA-G expression by human monocyte-derived dendritic cells. Mol Immunol. 2006 Jul;43(14):2151-60. Epub 2006 Feb 21. Pubmed: 16490253
- Teulings FA, Lems PH, Portengen H, Henkelman MS, Blonk DI: The action of 3-hydroxyanthranilic acid and other tryptophan metabolites on stimulated human lymphocytes. Acta Vitaminol Enzymol. 1975;29(1-6):113-6. Pubmed: 1244079
- Teulings FA, Mulder-Kooy GE, Peters HA, Fokkens W, Van Der Werf-Messing B: The excretion of 3-hydroxyanthranilic acid in patients with bladder and kidney carcinoma. Acta Vitaminol Enzymol. 1975;29(1-6):108-12. Pubmed: 1244078
- Werner ER, Lutz H, Fuchs D, Hausen A, Huber C, Niederwieser D, Pfleiderer W, Reibnegger G, Troppmair J, Wachter H: Identification of 3-hydroxyanthranilic acid in mixed lymphocyte cultures. Biol Chem Hoppe Seyler. 1985 Jan;366(1):99-102. Pubmed: 3159398
- Yeh JK, Brown RR: Effects of vitamin B-6 deficiency and tryptophan loading on urinary excretion of tryptophan metabolites in mammals. J Nutr. 1977 Feb;107(2):261-71. Pubmed: 833687
|
|---|
| Synthesis Reference: | Warnell, J. L. 3-Hydroxyanthranilic acid. Biochemical Preparations (1958), 6 20-4. |
|---|
| Material Safety Data Sheet (MSDS) | Download (PDF) |
|---|
| Links |
|---|
| External Links: | |
|---|