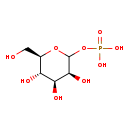Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
D-Mannose 1-phosphate (M2MDB000386)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-05-31 13:52:38 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 15:54:07 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | D-Mannose 1-phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | D-Mannose 1-phosphate is a normal metabolite intermediate in the Fructose and mannose metabolism, and substrate of phosphomannomutase 1 (PMM, EC: 5.4.2.8), an enzyme necessary for the synthesis of GDP-mannose. PMM converts mannose 6-phosphate to mannose-1-phosphate, which is required for the synthesis of GDP-mannose, a substrate for dolichol-linked oligosaccharide synthesis. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C6H13O9P | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 260.1358 Monoisotopic: 260.029718526 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | HXXFSFRBOHSIMQ-QTVWNMPRSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C6H13O9P/c7-1-2-3(8)4(9)5(10)6(14-2)15-16(11,12)13/h2-10H,1H2,(H2,11,12,13)/t2-,3-,4+,5+,6?/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 27251-84-9 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | {[(3S,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}phosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | D-mannose 1-phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | OC[C@H]1OC(OP(O)(O)=O)[C@@H](O)[C@@H](O)[C@@H]1O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as monosaccharide phosphates. These are monosaccharides comprising a phosphated group linked to the carbohydrate unit. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic oxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Carbohydrates and carbohydrate conjugates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Monosaccharide phosphates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic heteromonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | D-Mannose 1-phosphate <> Mannose 6-phosphate Guanosine diphosphate + Hydrogen ion + D-Mannose 1-phosphate > Guanosine diphosphate mannose + Phosphate Guanosine diphosphate mannose + Water > Guanosine monophosphate +2 Hydrogen ion + D-Mannose 1-phosphate Guanosine triphosphate + D-Mannose 1-phosphate <> Pyrophosphate + Guanosine diphosphate mannose Mannose 6-phosphate <> D-Mannose 1-phosphate Guanosine diphosphate mannose + Water > Hydrogen ion + Guanosine monophosphate + D-Mannose 1-phosphate Hydrogen ion + D-Mannose 1-phosphate + Guanosine triphosphate > Guanosine diphosphate mannose + Pyrophosphate α-D-mannose 6-phosphate > D-Mannose 1-phosphate D-Mannose 1-phosphate + Guanosine triphosphate + Hydrogen ion > Pyrophosphate + Guanosine diphosphate mannose Guanosine triphosphate + D-Mannose 1-phosphate <> Pyrophosphate + Guanosine diphosphate mannose Guanosine triphosphate + D-Mannose 1-phosphate <> Pyrophosphate + Guanosine diphosphate mannose | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in nucleotidyltransferase activity
- Specific function:
- Involved in the biosynthesis of the capsular polysaccharide colanic acid
- Gene Name:
- manC
- Uniprot ID:
- P24174
- Molecular weight:
- 53016
Reactions
| GTP + alpha-D-mannose 1-phosphate = diphosphate + GDP-mannose. |
- General function:
- Involved in intramolecular transferase activity, phosphotransferases
- Specific function:
- Involved in the biosynthesis of the capsular polysaccharide colanic acid
- Gene Name:
- manB
- Uniprot ID:
- P24175
- Molecular weight:
- 50462
Reactions
| Alpha-D-mannose 1-phosphate = D-mannose 6-phosphate. |
- General function:
- Involved in hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides
- Specific function:
- Catalyzes the hydrolysis of GDP-mannose. Can also use other substrates, catalyzing the hydrolysis of the pyrophosphate bond, releasing a nucleoside monophosphate and a phosphorylated moiety, depending on the substrate
- Gene Name:
- nudK
- Uniprot ID:
- P37128
- Molecular weight:
- 21749
Reactions
| GDP-mannose + H(2)O = GMP + mannose-1-phosphate. |