Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
Pantetheine 4'-phosphate (M2MDB000379)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-05-31 13:52:13 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 15:54:06 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Pantetheine 4'-phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Pantetheine 4'-phosphate is a metabolite in the pantothenate and coenzyme A biosynthesis pathway. It can be generated from pantatheine (via pantothenate kinase 1) or R-4'-phospho-pantothenoyl-L-cysteine (via phosphopantothenoylcysteine decarboxylase) or dephospho-CoA (via 4'-phosphopantetheine adenylyl-transferase and ectonucleotide pyrophosphatase). The conversion of pantetheine 4'-phosphate (4'-PP) to dephospho-CoA, is catalyzed by 4'-phosphopantetheine adenylyl-transferase. It has been identified as an essential cofactor in in the biosynthesis of fatty acids, polyketides, depsipeptides, peptides, and compounds derived from both carboxylic and amino acid precursors. In particular it is a key prosthetic group of acyl carrier protein (ACP) and peptidyl carrier proteins (PCP) and aryl carrier proteins (ArCP) derived from Coenzyme A. Phosphopantetheine fulfils two demands. Firstly, the intermediates remain covalently linked to the synthases (or synthetases) in an energy-rich thiol ester linkage. Secondly, the flexibility and length of phosphopantetheine chain (approximately 2 nm) allows the covalently tethered intermediates to have access to spatially distinct enzyme active sites. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
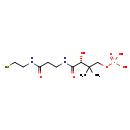
| Chemical Formula: | C11H23N2O7PS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 358.348 Monoisotopic: 358.096358302 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | JDMUPRLRUUMCTL-VIFPVBQESA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C11H23N2O7PS/c1-11(2,7-20-21(17,18)19)9(15)10(16)13-4-3-8(14)12-5-6-22/h9,15,22H,3-7H2,1-2H3,(H,12,14)(H,13,16)(H2,17,18,19)/t9-/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 2226-71-3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | [(3R)-3-hydroxy-2,2-dimethyl-3-({2-[(2-sulfanylethyl)carbamoyl]ethyl}carbamoyl)propoxy]phosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | 4'-phosphopantetheine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | CC(C)(COP(O)(O)=O)[C@@H](O)C(=O)NCCC(=O)NCCS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as beta amino acids and derivatives. These are amino acids having a (-NH2) group attached to the beta carbon atom. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Amino acids, peptides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Beta amino acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosine triphosphate + Hydrogen ion + Pantetheine 4'-phosphate <> Dephospho-CoA + Pyrophosphate 4-Phosphopantothenoylcysteine + Hydrogen ion <> Carbon dioxide + Pantetheine 4'-phosphate + pantotheine 4'-phosphate Acyl-carrier protein + Water + Acyl-carrier protein <> Pantetheine 4'-phosphate + Apo-[acyl-carrier-protein] Adenosine triphosphate + Pantetheine <> ADP + Pantetheine 4'-phosphate Adenosine triphosphate + Pantetheine 4'-phosphate <> Pyrophosphate + Dephospho-CoA 4-Phosphopantothenoylcysteine <> Pantetheine 4'-phosphate + Carbon dioxide Hydrogen ion + 4-Phosphopantothenoylcysteine > Pantetheine 4'-phosphate + Carbon dioxide Holo-[acyl-carrier-protein] + Water > Pantetheine 4'-phosphate + apo-[acyl-carrier-protein] Adenosine triphosphate + Hydrogen ion + Pantetheine 4'-phosphate <> Dephospho-CoA + Pyrophosphate Adenosine triphosphate + Hydrogen ion + Pantetheine 4'-phosphate <> Dephospho-CoA + Pyrophosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Masuda, Toru; Fujii, Shoichiro; Takanohashi, Kunio. Pantetheine 4'-phosphate. Jpn. Tokkyo Koho (1971), 3 pp. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in pantothenate kinase activity
- Specific function:
- ATP + (R)-pantothenate = ADP + (R)-4'- phosphopantothenate
- Gene Name:
- coaA
- Uniprot ID:
- P0A6I3
- Molecular weight:
- 36359
Reactions
| ATP + (R)-pantothenate = ADP + (R)-4'-phosphopantothenate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Reversibly transfers an adenylyl group from ATP to 4'- phosphopantetheine, yielding dephospho-CoA (dPCoA) and pyrophosphate
- Gene Name:
- coaD
- Uniprot ID:
- P0A6I6
- Molecular weight:
- 17837
Reactions
| ATP + pantetheine 4'-phosphate = diphosphate + 3'-dephospho-CoA. |
- General function:
- Involved in phosphopantothenate--cysteine ligase activity
- Specific function:
- Catalyzes two steps in the biosynthesis of coenzyme A. In the first step cysteine is conjugated to 4'-phosphopantothenate to form 4-phosphopantothenoylcysteine, in the latter compound is decarboxylated to form 4'-phosphopantotheine
- Gene Name:
- coaBC
- Uniprot ID:
- P0ABQ0
- Molecular weight:
- 43438
Reactions
| N-((R)-4'-phosphopantothenoyl)-L-cysteine = pantotheine 4'-phosphate + CO(2). |
| CTP + (R)-4'-phosphopantothenate + L-cysteine = CMP + diphosphate + N-((R)-4'-phosphopantothenoyl)-L-cysteine. |
- General function:
- Involved in [acyl-carrier-protein] phosphodiesterase activity
- Specific function:
- Converts holo-ACP to apo-ACP by hydrolytic cleavage of the phosphopantetheine prosthetic group from ACP
- Gene Name:
- acpH
- Uniprot ID:
- P21515
- Molecular weight:
- 22961
Reactions
| Holo-[acyl-carrier-protein] + H(2)O = 4'-phosphopantetheine + apo-[acyl-carrier-protein]. |
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for oligopeptides; probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- oppB
- Uniprot ID:
- P0AFH2
- Molecular weight:
- 33443
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for oligopeptides; probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- oppC
- Uniprot ID:
- P0AFH6
- Molecular weight:
- 33022
Transporters
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for oligopeptides; probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- oppB
- Uniprot ID:
- P0AFH2
- Molecular weight:
- 33443
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for oligopeptides; probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- oppC
- Uniprot ID:
- P0AFH6
- Molecular weight:
- 33022