| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 13:49:08 -0600 |
|---|
| Update Date | 2015-09-13 12:56:10 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
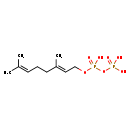
| Name: | Geranyl-PP |
|---|
| Description | Geranyl diphosphate is regarded as a key intermediate in the steroid, isoprene and terpene biosynthesis pathways and is used by organisms in the biosynthesis of farnesyl pyrophosphate, geranylgeranyl pyrophosphate, cholesterol, terpenes and terpenoids. (wikipedia). In E.coli, geranyl diphosphate synthase (GPPS) catalyzes the condensation of dimethylallyl diphosphate (DMAPP) and isopentenyl diphosphate (IPP) to form geranyl diphosphate. Geranyl pyrophosphate is an intermediate in the HMG-CoA reductase pathway used by organisms in the biosynthesis of terpenes and terpenoids. (Wikipedia) |
|---|
| Structure | |
|---|
| Synonyms: | - ω,E-geranyl diphosphate
- ω,e-geranyl diphosphoric acid
- (2E)-3,7-dimethylocta-2,6-dien-1-yl trihydrogen diphosphate
- (2E)-3,7-Dimethylocta-2,6-dien-1-yl trihydrogen diphosphoric acid
- Geranyl diphosphate
- Geranyl diphosphoric acid
- Geranyl pyrophosphate
- Geranyl pyrophosphoric acid
- Geranyl-diphosphate
- Geranyl-diphosphoric acid
- Geranyl-PP
- Geranyl-pyrophosphate
- Geranyl-pyrophosphoric acid
- GPP
- Monoterpenyl diphosphate
- Monoterpenyl diphosphoric acid
- Neryl diphosphate
- Neryl diphosphoric acid
- Omega,E-geranyl diphosphate
- Omega,e-geranyl diphosphoric acid
- Trans-Geranyl pyrophosphate
- trans-Geranyl pyrophosphoric acid
|
|---|
| Chemical Formula: | C10H20O7P2 |
|---|
| Weight: | Average: 314.2091
Monoisotopic: 314.068426018 |
|---|
| InChI Key: | GVVPGTZRZFNKDS-JXMROGBWSA-N |
|---|
| InChI: | InChI=1S/C10H20O7P2/c1-9(2)5-4-6-10(3)7-8-16-19(14,15)17-18(11,12)13/h5,7H,4,6,8H2,1-3H3,(H,14,15)(H2,11,12,13)/b10-7+ |
|---|
| CAS number: | 763-10-0 |
|---|
| IUPAC Name: | [({[(2E)-3,7-dimethylocta-2,6-dien-1-yl]oxy}(hydroxy)phosphoryl)oxy]phosphonic acid |
|---|
| Traditional IUPAC Name: | geranyl diphosphate |
|---|
| SMILES: | CC(C)=CCC\C(C)=C\CO[P@@](=O)(O)OP(=O)(O)O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as isoprenoid phosphates. These are prenol lipids containing a phosphate group linked to an isoprene (2-methylbuta-1,3-diene) unit. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Isoprenoid phosphates |
|---|
| Direct Parent | Isoprenoid phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Organic pyrophosphate
- Monoterpenoid
- Isoprenoid phosphate
- Acyclic monoterpenoid
- Monoalkyl phosphate
- Alkyl phosphate
- Phosphoric acid ester
- Organic phosphoric acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -2 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | | Secondary metabolites: methylerythritol phosphate and polyisoprenoid biosynthesis | PW000958 | 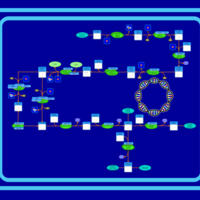 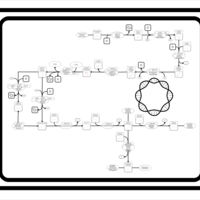  |
|
|---|
| KEGG Pathways: | - Metabolic pathways eco01100
- Terpenoid backbone biosynthesis ec00900
- Ubiquinone and other terpenoid-quinone biosynthesis ec00130
|
|---|
| EcoCyc Pathways: | - trans, trans-farnesyl diphosphate biosynthesis PWY-5123
|
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-004j-9640000000-9ce32c835bdcfa97fa15 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 6V, negative | splash10-0002-0090000000-b5ca864b0ba8818ad627 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 8V, negative | splash10-0002-0090000000-c7f7c5c529a56d532304 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 11V, negative | splash10-0002-3390000000-010bee66d1e4bd685e95 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 14V, negative | splash10-004i-9440000000-13e525f66230aae586a2 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 17V, negative | splash10-004i-9200000000-a9da8f03edb73eec50bf | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 20V, negative | splash10-004i-9100000000-fe3060700600ee72f8dd | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 23V, negative | splash10-004i-9000000000-15de26c097afd86b7007 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 27V, negative | splash10-004i-9000000000-bcc45bf0bf3bbb46af13 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 33V, negative | splash10-004i-9000000000-b8b56dc9ea06c2ce52b5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 21V, negative | splash10-0a4i-0900000000-f2685d197409ed952f02 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 21V, negative | splash10-004i-9000000000-5ef060d5ec4b80c35c1a | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 21V, negative | splash10-0a4i-0900000000-60b9e910670cbc6ebe0c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 21V, negative | splash10-0bt9-0390000000-6532032bbb594aa54d9c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 5V, negative | splash10-03di-0009000000-3bcab8c3688c96a4cec5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 7V, negative | splash10-03di-2009000000-42fa98db20ae4d6f687e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 9V, negative | splash10-01t9-9005000000-44618200345aed481b48 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 12V, negative | splash10-004i-9000000000-befed7a961a777eb5f66 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 15V, negative | splash10-004i-9000000000-8afaaf750b51acbd1a01 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 18V, negative | splash10-004i-9000000000-b5b3b346fecad6ae473d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00kr-1953000000-e5ee3340554c9ecac23e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-5910000000-82bcd2195a53b847981c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0gc0-9400000000-f87d391f1b3c01743515 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0509000000-43083a162145718cd977 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9500000000-053b6323e620a16328a8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-4faed4c921d605619caf | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Barnard GF, Popjak G: Human liver prenyltransferase and its characterization. Biochim Biophys Acta. 1981 Sep 15;661(1):87-99. Pubmed: 7295734
- Gan X, Kaplan R, Menke JG, MacNaul K, Chen Y, Sparrow CP, Zhou G, Wright SD, Cai TQ: Dual mechanisms of ABCA1 regulation by geranylgeranyl pyrophosphate. J Biol Chem. 2001 Dec 28;276(52):48702-8. Epub 2001 Oct 18. Pubmed: 11641412
- Holstein SA, Hohl RJ: Isoprenoids: remarkable diversity of form and function. Lipids. 2004 Apr;39(4):293-309. Pubmed: 15357017
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Kavanagh KL, Guo K, Dunford JE, Wu X, Knapp S, Ebetino FH, Rogers MJ, Russell RG, Oppermann U: The molecular mechanism of nitrogen-containing bisphosphonates as antiosteoporosis drugs. Proc Natl Acad Sci U S A. 2006 May 16;103(20):7829-34. Epub 2006 May 9. Pubmed: 16684881
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- Loza-Tavera H: Monoterpenes in essential oils. Biosynthesis and properties. Adv Exp Med Biol. 1999;464:49-62. Pubmed: 10335385
- Micali E, Chehade KA, Isaacs RJ, Andres DA, Spielmann HP: Protein farnesyltransferase isoprenoid substrate discrimination is dependent on isoprene double bonds and branched methyl groups. Biochemistry. 2001 Oct 16;40(41):12254-65. Pubmed: 11591144
- Pont F, Luciani B, Belmant C, Fournie JJ: Characterization of phosphoantigens by high-performance anion-exchange chromatography-electrospray ionization ion trap mass spectrometry and nanoelectrospray ionization ion trap mass spectrometry. Anal Chem. 2001 Aug 1;73(15):3562-9. Pubmed: 11510819
- Sagami H, Ogura K: [A new development in isoprenoid biochemistry brought by the discovery of prenylated proteins] Seikagaku. 1994 Dec;66(12):1488-501. Pubmed: 7884273
- Smit A, Mushegian A: Biosynthesis of isoprenoids via mevalonate in Archaea: the lost pathway. Genome Res. 2000 Oct;10(10):1468-84. Pubmed: 11042147
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
|
|---|
| Synthesis Reference: | Runquist M; Ericsson J; Thelin A; Chojnacki T; Dallner G Biosynthesis of trans,trans,trans-geranylgeranyl diphosphate by the cytosolic fraction from rat tissues. Biochemical and biophysical research communications (1992), 186(1), 157-65. |
|---|
| Material Safety Data Sheet (MSDS) | Download (PDF) |
|---|
| Links |
|---|
| External Links: | |
|---|