Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
Tetrahydropteridine (M2MDB000302)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-05-31 13:47:53 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 15:53:51 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Tetrahydropteridine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Tetrahydropteridine is a member of the chemical class known as Pteridines and Derivatives. These are polycyclic aromatic compounds containing a pteridine moiety, which consists of a pyrimidine fused to a pyrazine ring to form pyrimido(4,5-b)pyrazine. In a reversible reaction catalyzed by a nitroreductase / dihydropteridine reductase (EC 1.5.1.34), tetrahydropteridine can be converted to dihydropteridine while reducing NAD(P)+ to NAD(P)H. (KEGG) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C6H8N4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 136.1545 Monoisotopic: 136.074896276 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | IDAICLIJTRXNER-UHFFFAOYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C6H8N4/c1-2-9-6-5(8-1)3-7-4-10-6/h3-4,8H,1-2H2,(H,7,9,10) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 10593-78-9 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
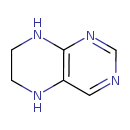
| IUPAC Name: | 5,6,7,8-tetrahydropteridine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | 5,6,7,8-tetrahydropteridine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | C1CNC2=C(N1)C=NC=N2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as pteridines and derivatives. These are polycyclic aromatic compounds containing a pteridine moiety, which consists of a pyrimidine fused to a pyrazine ring to form pyrimido(4,5-b)pyrazine. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organoheterocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Pteridines and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Pteridines and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | 6,7-Dihydropteridine + 3 Hydrogen ion + NADH <> NAD + Tetrahydropteridine 6,7-Dihydropteridine + 3 Hydrogen ion + NADPH <> NADP + Tetrahydropteridine NAD(P)<sup>+</sup> + Tetrahydropteridine <> NAD(P)H + 6,7-Dihydropteridine + Hydrogen ion Tetrahydropteridine + NAD > 6,7-Dihydropteridine + NADH Tetrahydropteridine + NADP > 6,7-Dihydropteridine + NADPH Tetrahydropteridine + NAD + NADP <> 6,7-Dihydropteridine + NADH + NADPH + Hydrogen ion | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in oxidoreductase activity
- Specific function:
- Various electron acceptors are also reduced by HMP in vitro, including dihydropterine, ferrisiderophores, ferric citrate, cytochrome c, nitrite, S-nitrosoglutathione, and alkylhydroperoxides. However, it is unknown if these reactions are of any biological significance in vivo
- Gene Name:
- hmp
- Uniprot ID:
- P24232
- Molecular weight:
- 43867
Reactions
| 2 NO + 2 O(2) + NAD(P)H = 2 NO(3)(-) + NAD(P)(+). |
- General function:
- Involved in oxidoreductase activity
- Specific function:
- Reduction of a variety of nitroaromatic compounds using NADH (and to lesser extent NADPH) as source of reducing equivalents; two electrons are transferred. Capable of reducing nitrofurazone, quinones and the anti-tumor agent CB1954 (5- (aziridin-1-yl)-2,4-dinitrobenzamide). The reduction of CB1954 results in the generation of cytotoxic species
- Gene Name:
- nfnB
- Uniprot ID:
- P38489
- Molecular weight:
- 23905
Reactions
| A 5,6,7,8-tetrahydropteridine + NAD(P)(+) = a 6,7-dihydropteridine + NAD(P)H. |