| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 13:44:57 -0600 |
|---|
| Update Date | 2015-06-03 15:53:43 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
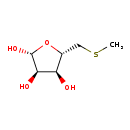
| Name: | 5-Methylthioribose |
|---|
| Description | S-methyl-5-thio-D-ribose is a product of the enzyme 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase. This enzyme catalyzes the irreversible cleavage of the glycosidic bond in 5'-methylthioadenosine (MTA) to produce adenine and the corresponding thioribose, 5'-methylthioribose. |
|---|
| Structure | |
|---|
| Synonyms: | - 5-Deoxy-5-(methylthio)ribose
- 5-Methylthio-D-ribose
- 5-Methylthioribose
- S-methyl-5-thio-D-ribofuranose
- MTR
- S(5)-Methyl-5-thio-D-ribose
- S-Methyl-5-thio-D-ribofuranose
- S5-Methyl-5-thio-D-ribose
|
|---|
| Chemical Formula: | C6H12O4S |
|---|
| Weight: | Average: 180.222
Monoisotopic: 180.045629562 |
|---|
| InChI Key: | OLVVOVIFTBSBBH-KVTDHHQDSA-N |
|---|
| InChI: | InChI=1S/C6H12O4S/c1-11-2-3-4(7)5(8)6(9)10-3/h3-9H,2H2,1H3/t3-,4-,5-,6-/m1/s1 |
|---|
| CAS number: | 23656-67-9 |
|---|
| IUPAC Name: | (2R,3R,4S,5S)-5-[(methylsulfanyl)methyl]oxolane-2,3,4-triol |
|---|
| Traditional IUPAC Name: | 5-methylthio-D-ribose |
|---|
| SMILES: | CSC[C@H]1O[C@@H](O)[C@H](O)[C@@H]1O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pentoses. These are monosaccharides in which the carbohydrate moiety contains five carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Pentoses |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pentose monosaccharide
- Tetrahydrofuran
- Secondary alcohol
- Hemiacetal
- 1,2-diol
- Oxacycle
- Organoheterocyclic compound
- Dialkylthioether
- Sulfenyl compound
- Thioether
- Polyol
- Hydrocarbon derivative
- Organosulfur compound
- Alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | 0 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | | S-adenosyl-L-methionine biosynthesis | PW000837 |    | | Spermidine biosynthesis and metabolism | PW002085 | 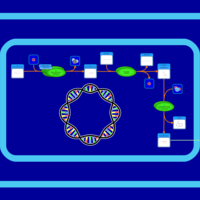 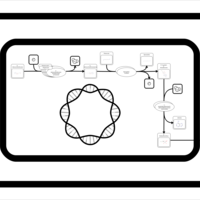 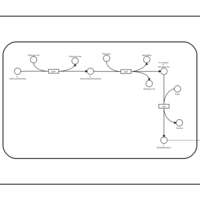 |
|
|---|
| KEGG Pathways: | - Cysteine and methionine metabolism ec00270
|
|---|
| EcoCyc Pathways: | - S-methyl-5'-thioadenosine degradation IV PWY0-1391
|
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0nor-9600000000-757c4b3ed6add1bdc168 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-0719-9247000000-321d54ddf2f38682b57c | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0900000000-7914d8991b0a6af1d99f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-02ai-2900000000-e36191d55523024bf643 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0bvj-9500000000-94cbb1e527c7c7e21314 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-9200000000-21073f76322842d53a46 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-9300000000-89d811f24fdb17e739e8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0002-9000000000-ed328bd35b3fe1440450 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-1900000000-aefd2630eb3d70da8200 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-9100000000-57ad8e0e93b83006dc3c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0002-9000000000-789718691297aaebb3ad | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01qa-4900000000-1a70ce18540d6a0916d8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-8900000000-ca12a9fcff10da622383 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03dj-9000000000-e18a37fac06996f6a660 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Carteni-Farina M, della Ragione F, Cacciapuoti G, Porcelli M, Zappia V: Transport and metabolism of 5'-methylthioadenosine in human erythrocytes. Biochim Biophys Acta. 1983 Jan 19;727(2):221-9. Pubmed: 6838867
- Della Ragione F, Carteni-Farina M, Gragnaniello V, Schettino MI, Zappia V: Purification and characterization of 5'-deoxy-5'-methylthioadenosine phosphorylase from human placenta. J Biol Chem. 1986 Sep 15;261(26):12324-9. Pubmed: 3091600
- Gianotti AJ, Tower PA, Sheley JH, Conte PA, Spiro C, Ferro AJ, Fitchen JH, Riscoe MK: Selective killing of Klebsiella pneumoniae by 5-trifluoromethylthioribose. Chemotherapeutic exploitation of the enzyme 5-methylthioribose kinase. J Biol Chem. 1990 Jan 15;265(2):831-7. Pubmed: 2153115
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|